Integrative Hi-C, ATAC-seq And ChIP-seq Analysis Provide Insights Into Gene Regulation Mechanisms
Integrative analysis of multiple genomic datasets is crucial for understanding the complexity of gene regulation in health and disease. Recent advances in high-throughput sequencing technologies have led to the generation of large amounts of genomic data, such as Hi-C, ATAC-seq, ChIP-seq, and gene expression data. Hi-C is a technique that captures chromatin interactions genome-wide, while ATAC-seq and ChIP-seq provide information on chromatin accessibility and protein-DNA interactions, respectively. By integrating these datasets, researchers can investigate the relationship between chromatin structure, transcription factor binding, and gene expression.
Dissecting the mechanism by which Runx1 affects gene expression in hematopoietic development through dynamic chromatin boundaries
Runx1/AML1 is a member of the RUNX family of transcription factors involved in the maintenance of normal hematopoietic homeostasis. Disruption of RUNX1 in humans leads to several hematopoietic disorders, including acute myeloid leukemia and familial platelet disorders with myeloid malignancies. Runx1 is a large and complex gene that is transcriptionally regulated by two promoters (P1 and P2) under the spatiotemporal control of multiple hematopoietic enhancers. Elucidation of the transcriptional regulatory mechanism of Runx1 is expected to contribute to a better understanding of the chromatin conformational changes adopted by complex multi-promoter genes during development. Combined analysis of Dnasel-seq, ChIP-seq, ATAC-seq and RNA-seq revealed accessible chromatin sites on known enhancers, CTCF sites and candidate cis-regulatory elements within TAD during differentiation. It was found that during differentiation, sub-TAD chromatin boundaries are dynamically formed within the large and complex Runx1 regulatory domain and are involved in coordinating gene expression and hematopoietic differentiation.
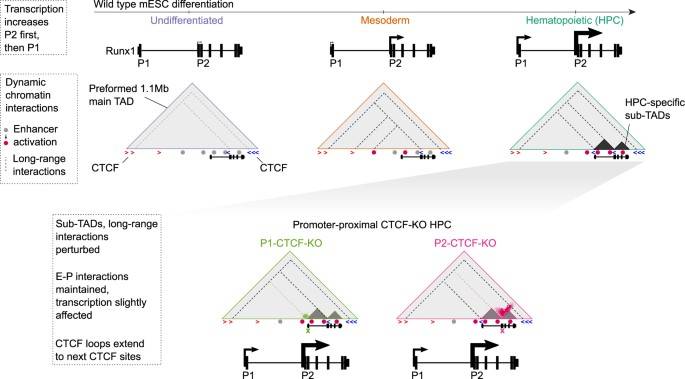 Schematic model of dynamic chromatin changes at Runx1 during hematopoietic development and after promoter-proximal CTCF site deletions.
Schematic model of dynamic chromatin changes at Runx1 during hematopoietic development and after promoter-proximal CTCF site deletions.
Dissecting long-range transcriptional cis-regulatory elements in allotetraploid cotton (Gossypium hirsutum) genome
In addition to disease research, integrative analysis has also been used to study normal development and cellular differentiation. Gossypium hirsutum, an imported fiber and oilseed crop widely grown worldwide, is also a model for analyzing all polyploidized genomes. This research focuses on determining the functional genome composition, including protein-coding genes and non-coding regulatory elements, to guide the improvement of agricultural traits. The study first prepared an ultra-high resolution (3 Kb) Hi-C library (233 Gb) and a Pore-C library with appropriate resolution (20 Kb) based on leaves of G. hirsutum to construct a fine 3D genome map of cotton to fully explore the multiple interactions of chromatin structural domains to help understand the state and intranuclear location of functional genomic components. Next, Hi-C, ChIA-PET (H3K4me3) and ChIP-Seq (H3K4me3, H3K27ac and H3K27me3) were used to investigate chromatin loops and their potential role in gene transcription. At a resolution of 3 Kb, the study observed 31047 gene-gene loops, 40035 gene-non-coding region loops and 121,415 other loops. Genes located at gene-gene loop anchor points had higher expression levels, which were associated with higher levels of active histone modifications. Possible long-range transcriptional cis-regulatory elements (CREs) were subsequently identified by combining ATAC-Seq, ChIP-Seq and chromatin loops. This study comprehensively probed the fine 3D genome structure of allotetraploid cotton and analyzed the characteristics of large and complex high-level multiple interactions and the potential impact on gene expression.
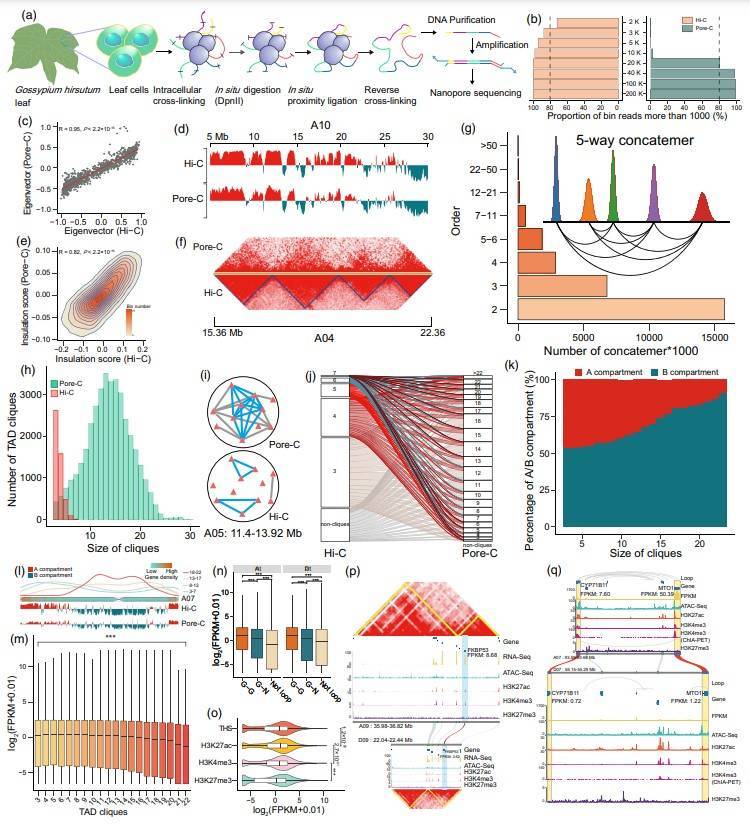 Multi-omics mapping of the fine 3D genome structure and implications in transcriptional regulation in allotetraploid cotton.
Multi-omics mapping of the fine 3D genome structure and implications in transcriptional regulation in allotetraploid cotton.
Integrative analysis of multiple genomic datasets, such as Hi-C, ATAC-seq, ChIP-seq, and gene expression data, has the potential to provide a deeper understanding of the spatial organization of chromatin, transcriptional regulation, and disease mechanisms.
References:
- Owens D D G, Anselmi G, Oudelaar A M, et al. Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development. Nature communications, 2022, 13(1): 1-15.
- Huang X, Tian X, Pei L, et al. Multi-omics mapping of chromatin interaction resolves the fine hierarchy of 3D genome in allotetraploid cotton. Plant biotechnology journal, 2022.


 Sample Submission Guidelines
Sample Submission Guidelines
