Immune Repertoire Sequencing Accelerates Vaccine Development
With the emergence of antibiotics, the pandemic or local epidemic caused by pathogenic bacteria has declined significantly. However, the abuse of antibiotics has accelerated the emergence of drug-resistant bacteria and the number of deaths due to drug-resistant bacteria in the world is increasing year by year, microbial drug resistance has become an increasingly serious global crisis, which bring about a great threat to human health. New methods that can replace antibiotic treatment are urgently needed in addition to regulating the use of antibiotics and accelerating the development of new antibiotics, vaccines are considered to be an effective tool to mitigate the drug resistance crisis.
Due to the next-generation sequencing technology is developing vigorously, and the increasing application of immunization in disease diagnosis and treatment, the sequencing of the Immune repertoire has been widely studied. Immune repertoire sequencing refers to take T/B lymphocytes as the research target and use multiple PCR technology to amplify the complementary determinant region (CDR3 region) that determines the diversity of B cell receptor (BCR) or T cell receptor (TCR), and then combining high-throughput sequencing technology to comprehensively evaluate the diversity of the immune system. At present, the technology of high-throughput sequencing of the immunoglobulin library has been applied to the research of many diseases, including vaccine design and evaluation, biomarkers for tumor diagnosis, classification and prognosis, detection of minimal residual disease (MRD), autoimmune diseases, tumor research and post transplantation monitoring.
Immune repertoire sequencing facilitates the study of antibodies induced by COVID-19 vaccine
The COVID-19 has continued since the end of 2019, which posed great challenges to global public health security and resulted in economic crisis worldwide. Among the approved vaccines and vaccine candidates in clinical evaluation, except inactivated virus vaccines, the others make use of SARS-CoV-2 receptor binding domain (RBD) or spike protein (S) as vaccine immunogen. However, few studies have systematically compared the antibody libraries induced by RBD and S immunogens. Researchers employed 10×Genomics Immune Repertoire sequencing to yield important insights into antibody responses to different SARS-CoV-2 antigens. The results indicated that in contrast to RBD, the S immunogen elicited diverse neutralizing mAbs that targeted not only RBD but also NTD. Those diverse mAbs have potential to exert potent neutralizing activity against SARS-CoV-2 through synergistic effect and are generally beneficial to combat against virus variants. Most importantly, these different antibodies may resist the risk of vaccine failure caused by virus mutation through synergy to a certain extent.
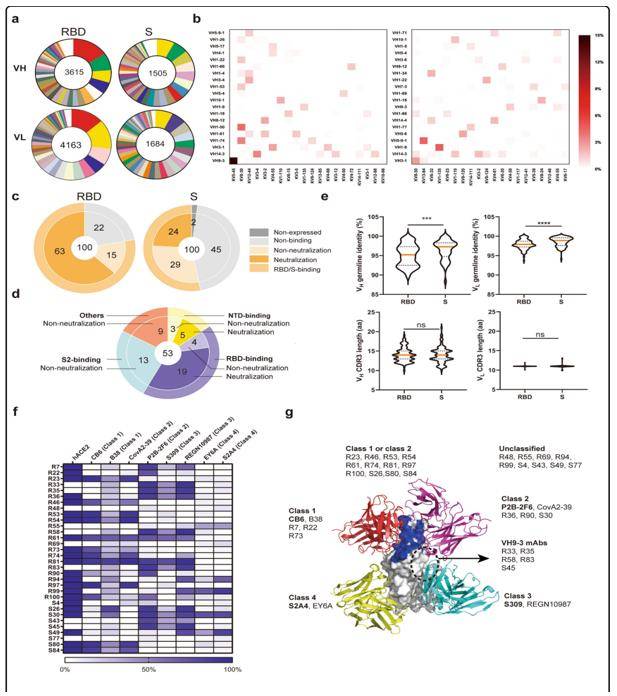 Distinct BCR repertoires elicited by SARS-CoV-2 RBD and S vaccinations in mice
Distinct BCR repertoires elicited by SARS-CoV-2 RBD and S vaccinations in mice
The application of B cells sequencing in convalescent patients
Recent report shows the rapid and efficient identification of SARS-CoV-2-neutralizing antibodies achieved by high-throughput single-cell RNA and VDJ sequencing of antigen-binding B cells from convalescent COVID-19 patients. Over 8,500 antigen-binding B cell clonotypes expressing immunoglobulin G1 (IgG1) antibodies were identified from 60 convalescent patients. BCR sequencing could lead to the identification of highly potent neutralizing mAbs that have strong therapeutic and prophylactic efficacy and it is of great significance to the prognosis of disease and vaccine design, which could greatly assist in the intervention of prevailing and emerging infectious diseases, such as COVID-19.
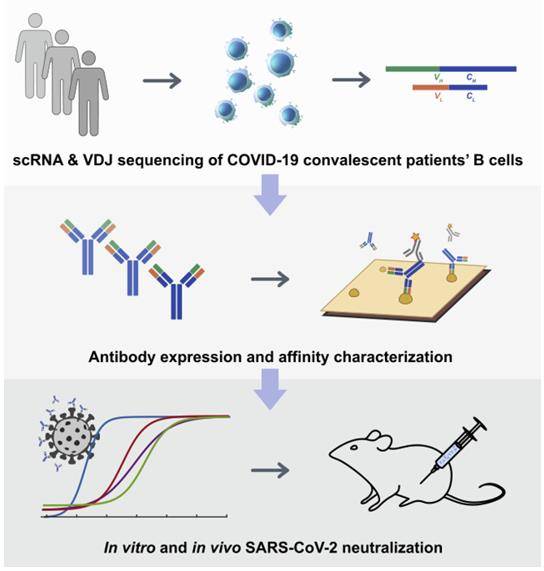 High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells
High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells
Immune repertoire sequencing has matured into a powerful technology for antigen discovery. More studies are expected in the years to come to shed light on burning questions in the field. Discovered antigens by this method can be directly encoded in nucleic acid-based vaccines, constituting a versatile and powerful vaccine development pipeline. In the future, Immune Repertoire Sequencing technology can not only promote the rapid development of bacterial vaccines (especially vaccines targeting intracellular bacteria), but also make greater efforts in the discovery of new tumor antigens, assisting mRNA vaccines move towards more sophisticated development. Further improvements are expected to boost the number of identified antigens and to strengthen the golden alliance with viral vector and mRNA vaccine platforms, facilitating development of effective, novel antibacterial vaccines.
References:
- Lavinder JJ, Wine Y, Giesecke C, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci U S A. 2014;111(6):2259-2264.
- Sureshchandra, Suhas et al. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI insight vol. 6,24 e153201. 22 Dec. 2021.
- Tian, Siyu et al. Distinct BCR repertoires elicited by SARS-CoV-2 RBD and S vaccinations in mice. Cell discovery vol. 7,1 91. 7 Oct. 2021.
- Cao, Yunlong et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells. Cell vol. 182,1 (2020): 73-84.e16.


 Sample Submission Guidelines
Sample Submission Guidelines
