Dual RNA Sequencing: Definition & Principle, Workflow, and Applications
What is dual RNA sequencing?
Understanding how pathogen induces disease requires the knowledge on which genes are expressed and how they are regulated during infection. RNA sequencing has been routinely used to analyze the gene expression of microbial pathogens. Apart from the standard mRNA sequencing, new methods are surging based on RNA-seq, mapping the post-transcriptional networks controlled by small RNAs and to discover associated RNA-binding proteins in the pathogen itself. Dual RNA sequencing is one of the novel RNA-seq technologies. It shines a light on the simultaneous transcriptomics in host-pathogen interactions and is particularly suitable for investigating the molecular mechanisms of hosts and pathogens interactions. Dual RNA-seq does not require predesigned species-specific probes and allows the detection of transcripts with low abundances in a sensitive manner. In addition, it enables the analysis of specific transcripts for pathogen and host in mixed samples from at least two species, revealing the role of non-coding RNA in the process of infection. The correlation between the microbial gene activity and specific host reactions can also be clarified. Dual RNA sequencing can facilitate the studies of various fields including pathology and disease treatment.
The workflow of dual RNA sequencing
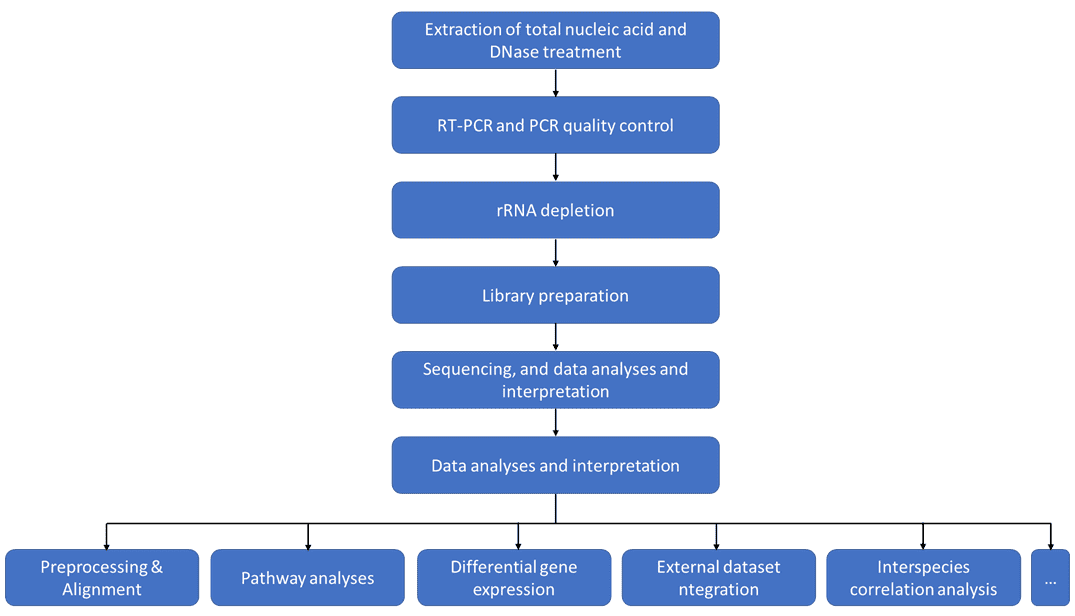
Dual RNA-Seq is the parallel transcriptomic analysis of bacterial pathogens and their eukaryotic host cells, which generally involves a typical workflow including RNA extraction, rRNA depletion, library preparation, sequencing, and data analyses and interpretation. Meanwhile, the data analysis procedures include preprocessing, alignment, and further advanced downstream analyses such as alternative and non-linear splicing, differential expression, epigenetic analyses, which finally deliver an outcome covering quality reports, calculations and predictions for novel transcripts, probabilities of differentially expressed transcripts and transcript characterizations.
Similar to conventional transcriptomic methodologies, dual RNA-seq involves quantification, screening of differences, pathway annotation, network construction, and so on. With regard to the earlier part of the process, the corresponding reference genome is needed for the following bioinformatics analyses.
Applications
Dual RNA-Seq addresses the interaction between pathogens and their hosts, where total RNA is extracted from infected cells. The sequencing and analyses of the interested RNAs from bacterial pathogens and infected cells can be followed at the same time, while the mixed sequencing reads are assigned to their originating genomes. Dual RNA-Seq is based on the improvement of library construction and sequencing technology. It can facilitate separating research into multiple directions including but not limited to:
- Gene expression level variations of host and pathogen
- Studying infection-related sRNA, mitochondrial RNA, etc.
- In-depth study of signal path-related noncoding RNA
- Studying the differences between wild and mutant pathogens via comparative genomics
- Uncovering the interaction mechanism between/among species by studying the gene regulation relationship(s)
- Studying the regulatory network, pathogenic mechanism during infection, and host resistance mechanism in the pathogen-host interaction process
- Studying the evolutionary relationships among pathogens of different species, and further discovery of positive selection based on homologous genes
References:
- Westermann A; et al. Resolving host–pathogen interactions by dual RNA-seq. PLOS Pathogens. 2017, 13.
- Marsh JW; et al. A Laboratory Methodology for Dual RNA-Sequencing of Bacteria and their Host Cells In Vitro. Frontiers in microbiology. 2017, 8.


 Sample Submission Guidelines
Sample Submission Guidelines
