Transposon Dynamics in Drosophila simulans Populations
Transposons, also known as transposable elements, play a significant role in shaping the genetic landscapes of various organisms. In the case of the fruit fly Drosophila melanogaster, the genomic distribution and activity of transposons have been meticulously documented. Studies have estimated that the rate of transposon insertions in D. melanogaster hovers around X insertions per copy per generation. However, these insertions often remain at low frequencies within the population due to the selective pressures of natural selection or because they are in the early stages of a transpositional event.
Global Dispersal and Transposon Dynamics
Drosophila melanogaster, originally native to Africa, has spread worldwide in tandem with human migrations. Yet, as this species ventures into new environments, it encounters novel selective pressures. Consequently, these encounters can awaken dormant transposons in the genome, leading to heightened transposition activity. As a result, Drosophila melanogaster populations outside of Africa typically exhibit a higher prevalence of transposons compared to their African counterparts.
Comparative Insights from Drosophila simulans
A closely related species, Drosophila simulans, also originated in the African tropics but has undergone global dispersal, albeit more recently over the last century. Studies have revealed significant disparities in transposon content among Drosophila simulans populations, raising the possibility that transposons in this species might be reawakening and initiating transpositional bursts.
Methodological Approaches
Numerous investigations comparing transposons in Drosophila melanogaster and Drosophila simulans have relied on pooled sequencing, or pool-seq. Through these studies, it has been established that transposon content in Drosophila melanogaster is lower in African populations compared to those from other regions. Additionally, it has been observed that P elements from Drosophila melanogaster have invaded the Drosophila genome.
This paper presents a comprehensive examination of transposon dynamics in Drosophila melanogaster, drawing comparative insights from Drosophila simulans populations.
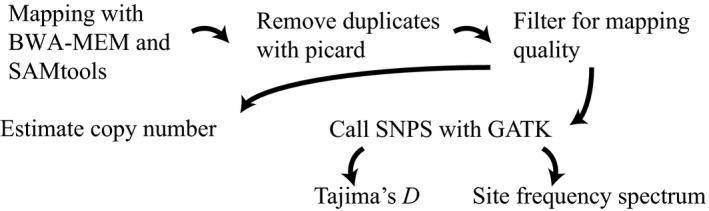 An outline of the pipeline used for estimating copy number of transposable elements in Drosophila simulans, as well as estimating the site frequency spectrum. (Signor et al., 2020)
An outline of the pipeline used for estimating copy number of transposable elements in Drosophila simulans, as well as estimating the site frequency spectrum. (Signor et al., 2020)
Comparative Analysis of Drosophila Populations from Africa and the Americas
This investigation involved the comparison of five natural Drosophila strains from Africa, sixteen laboratory strains of Drosophila, and one hundred and sixty-nine laboratory strains of Drosophila originating from the Americas.
Out of a total of 128 transposons, 85 exhibited variations in copy numbers between the African and American populations. The majority of these displayed higher copy numbers within the American population, with only 17 transposons showing greater prevalence in the African population. The five transposons with the most significant disparities between these two populations were identified as INE-1, Tc1, transib2, 1360, and Cr1a.
In concurrence with prior research findings, the presence of pogo and Helitron transposons was not detected in Drosophila simulans. A study utilizing pooled sequencing in 2015 reported that the most abundant transposons in Drosophila included INE-1, roo, Cr1a, Rt1c, and hobo, while G6 exhibited the lowest prevalence. There are notable resemblances between these outcomes and the results presented in this study.
This study also involved a comparison of the locus spectrum of transposons in African and American populations of Drosophila simulans. Notably, the American population was found to harbor a higher number of medium-frequency transposons in accordance with results obtained from Tajima's D analysis. As for G6, flea, Bari1, and Juan, these transposons displayed a marked bias towards lower frequencies in both populations, indicating that they might be currently active transposons. Furthermore, other studies have suggested that Juan is actively transposing in Drosophila melanogaster.
The P-element, according to studies, appears to have migrated from the Drosophila melanogaster genome into the Drosophila melanogaster genome.
Comparing Natural and Laboratory Strains
In general, laboratory strains undergo inbreeding, which typically has minimal effects on transposon copy numbers. In the context of this study, no significant differences were observed in transposon copy numbers between laboratory and natural strains within the African population.
Dmau\mariner also exhibited higher copy numbers in the African population than in the American populations, with nearly no observed polymorphisms. This suggests that this particular transposon is experiencing rapid and widespread dissemination within Drosophila simulans, consistent with prior research findings.
It's worth noting that the number of G6 transposon copies identified in this study exceeded previous literature reports. Nevertheless, considering that G6 was predominantly present at low frequencies in this study, it suggests that the transposon is also undergoing extensive diffusion.
Comparison with Drosophila melanogaster
In Drosophila melanogaster, the transposons with the highest copy numbers are INE-1, 1360, jockey, hobo, roo, and the P-element. Conversely, in the two Drosophila populations under study, the transposons with the highest copy numbers are G6, Cr1a, and INE-1.
Remarkably, in both Drosophila melanogaster and Drosophila simulans, African populations exhibited lower copy numbers compared to populations from other regions. Therefore, the global dispersion of both species was accompanied by an increase in transpositional activity.
This study's findings indicate that the following transposons are active in Drosophila melanogaster: Dsim\ninja, Dmau\mariner, P-element, gypsy10, opus, blood, GATE, diver, Tabor, INE-1, diver2, Idefix, 1731, 412, 297, G6, flea, Bari1, Transpac, Tabor, accord, and Juan.
Reference:
- Signor, Sarah. "Transposable elements in individual genotypes of Drosophila simulans." Ecology and Evolution 10.7 (2020): 3402-3412.


 Sample Submission Guidelines
Sample Submission Guidelines
