Strand-Specific RNA-Seq Used to Study Gene Function and Pathogenic Mechanisms
Strand-specific RNA-seq means that the direction information of RNA chain is saved in the sequencing library during library construction, and the data analysis after sequencing can determine whether the transcript is from the sense chain or the antisense chain. Compared with ordinary transcriptome sequencing, it can more accurately count the number of transcripts and determine the structure of genes, and can find more antisense transcripts, which is currently widely used in the research of gene structure, gene expression regulation and other fields.
Advantages and application of strand-specific RNA-seq:
- Identify antisense transcripts
- Annotate a genome
- Discover novel transcripts and structural analysis of model organism
- Obtain more accurate quantification of gene expression and predict new transcripts
- Improve the accuracy of discovering circRNA
Strand-specific RNA-seq helps to explore the mysteries of RNA editing mechanisms
RNA editing is a kind of post transcriptional regulation mechanism of genes, which refers to the insertion, deletion or replacement of bases on RNA molecules, leading to the difference between mature RNA and the DNA template that originally encoded it. RNA editing systems have arisen multiple times within eukaryotes and involve a range of posttranscriptional processing mechanisms that alter RNA sequences by the insertion, deletion, or substitution of nucleotides but exclude splicing, 5' -capping, and 3' -polyadenylation by convention. Extensive adenosine-to-inosine (A-to-I) editing of nuclear-transcribed mRNAs is the hallmark of metazoan transcriptional regulation. Here, the researchers elaborately selected 22 species that can represent different stages of animal evolution, and conducted high depth genome re sequencing and chain specific transcriptome sequencing for each species. Subsequently, RNA editing maps of these species were constructed using the team's self-developed RES Scanner software package. They provide substantial evidence supporting A-to-I mRNA editing as a regulatory innovation originating in the last common ancestor of extant metazoans. The sequencing results showed that A-to-I editing mainly occurred in the non-coding RNA transcribed by the genome repeat sequence, especially the young repeat sequence, while editing events occurred in the protein coding region were very rare and intermolecular pairing of sense-antisense transcripts as an important mechanism for forming dsRNA substrates for A-to-I editing in some but not all lineages. In general, the metazoan A-to-I editing may have evolved initially as a defense mechanism against repeat-derived dsRNA, but due to its mutagenic character, it was eventually incorporated into a variety of biological processes.
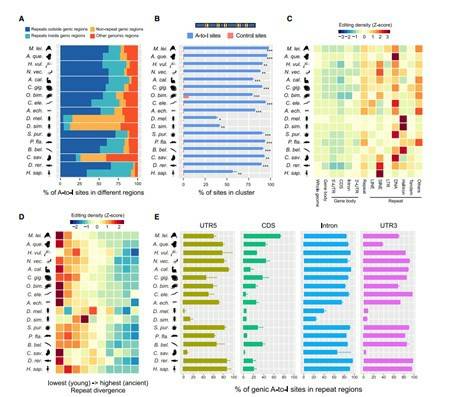 The genomic targets of metazoan A-to-I editing
The genomic targets of metazoan A-to-I editing
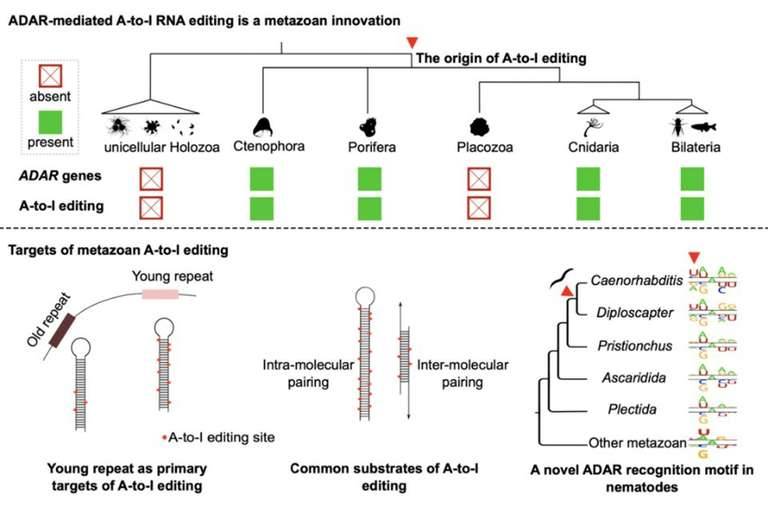 The Origin and Main Substrates of RNA Editing Mechanism Mediated by ADAR
The Origin and Main Substrates of RNA Editing Mechanism Mediated by ADAR
Cotton is an important natural fiber crop. Recently, researchers integrate four complementary high-throughput techniques, including PacBio Iso-seq, strand-specific RNA-seq, CAGE-seq, and PolyA-seq, to systematically explore the transcription landscape across 16 tissues or different organ types in Gossypium arboreum. They designed a data analysis process called IGIA to integrate and reconstruct accurate gene structure annotation information from different sequencing techniques. The results revealed a dynamic and diverse transcriptome map in cotton, including tissue specific gene expression, selective use of transcription start sites (TSS) and polyadenylation sites, hot spots of alternative splicing, and transcriptional read through. These regulatory events will affect many genes in various aspects, such as the acquisition or loss of functional RNA motif and protein structure domains, fine-tuning of DNA binding activity, and co-regulation of genes in the same complex or pathway. These methods and discoveries provide valuable resources for further functional genome research, such as understanding the natural SNP variation in plant populations.
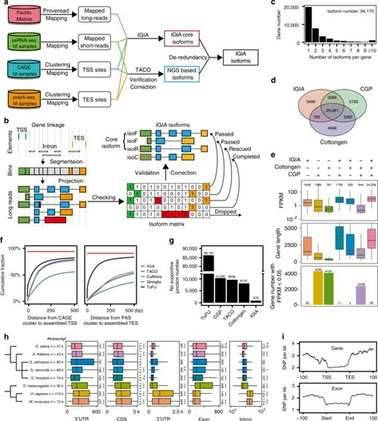 Integrative multi-strategic RNA-seq for the high-resolution RNA landscape in G. arboreum
Integrative multi-strategic RNA-seq for the high-resolution RNA landscape in G. arboreum
The strand-specific RNA-seq has great advantages. It is believed that with the decrease of the overall cost of the sequencing industry, the experimental demand for chain specific transcriptome will become greater and greater, which provides a good help for the research of RNA related topics such as transcriptome differential gene expression and splicing variants.
References:
- Zhang, Pei et al. “On the origin and evolution of RNA editing in metazoans." Cell reports, vol. 42,2 112112. 14 Feb. 2023
- Wang, Kun et al. "Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton." Nature communications vol. 10,1 4714. 17 Oct. 2019


 Sample Submission Guidelines
Sample Submission Guidelines
