Mapping Early Cancer Mutations with Single-Molecule cfDNA Sequencing
The effectiveness of cancer treatment and the duration of survival are intricately linked with timely cancer detection, underscoring the significance of early diagnosis. However, the current methods employed for clinical cancer screening present notable challenges. For instance, while LDCT screening has demonstrated a capacity to reduce mortality, its adoption among high-risk populations remains limited, partially due to potential drawbacks such as low specificity, exposure to radiation, and instances of overdiagnosis. Furthermore, certain cancer types still lack a suitable screening approach, even though prompt detection holds the potential to enhance patient prognosis.
A potential solution to these challenges lies in liquid biopsy, which identifies cancer through circulating tumor DNA (ctDNA) within cell-free DNA (cfDNA). Nevertheless, cfDNA typically carries a modest proportion of mutational data within the cancer genome, posing difficulties in distinguishing genuine mutational signals from the backdrop of sequencing noise.
Highlights
The group of researchers has formulated an approach for detecting numerous somatic mutations in cfDNA through a technique called GEMINI (GEnome-wide Mutational Incidence for Non-Invasive detection of cancer) using single-molecule sequencing. Employing GEMINI, they unveiled distinct mutation type frequencies in various tissue and cfDNA regions among cancer patients. These frequencies were found to be linked with replication time and other chromatin characteristics. Moreover, the comprehensive mutation spectrum identified by GEMINI, combined with additional attributes and a machine learning model, enabled the identification of over 90% of lung cancer cases, encompassing stage I and II patients.
 Approach for cancer detection using single-molecule cfDNA sequencing. (Bruhm et al., 2023)
Approach for cancer detection using single-molecule cfDNA sequencing. (Bruhm et al., 2023)
Utilizing Somatic Mutations for Non-Invasive Cancer Diagnosis
In a comprehensive study, scientists scrutinized genomic sequences of cancer from 2,511 individuals representing 25 distinct cancer types as part of the Pan-Cancer Analysis of the Whole Genome (PCAWG) project. Their investigation unveiled varying mutation frequencies in the genomes of different tumor categories. For instance, lung cancer exhibited an average of 52,209 somatic mutations per genome. The team employed an approach wherein 31 matched PCAWG samples were examined, segmenting the sequence data, encompassing 3,076,901 mutations, into 1,144 non-overlapping 2.5 Mb sections. Remarkably, numerous tumors exhibited shared regions with heightened mutation frequencies across their genomic landscapes.
To assess the efficacy of GEMINI in detecting tumor-derived DNA, the researchers pinpointed genomic areas showcasing the highest occurrence of C>A mutations within a training set of both cancers and control samples. They observed that regions enriched for C>A changes were present in the 31 PCAWGs but not in normal samples, suggesting that isolating mutation frequencies between cancer and control regions could effectively eliminate background mutations in a given patient's sample. By utilizing the C>A region frequency, GEMINI successfully differentiated between cancer and non-cancer samples with remarkable precision, achieving an impressive AUC of 0.91.
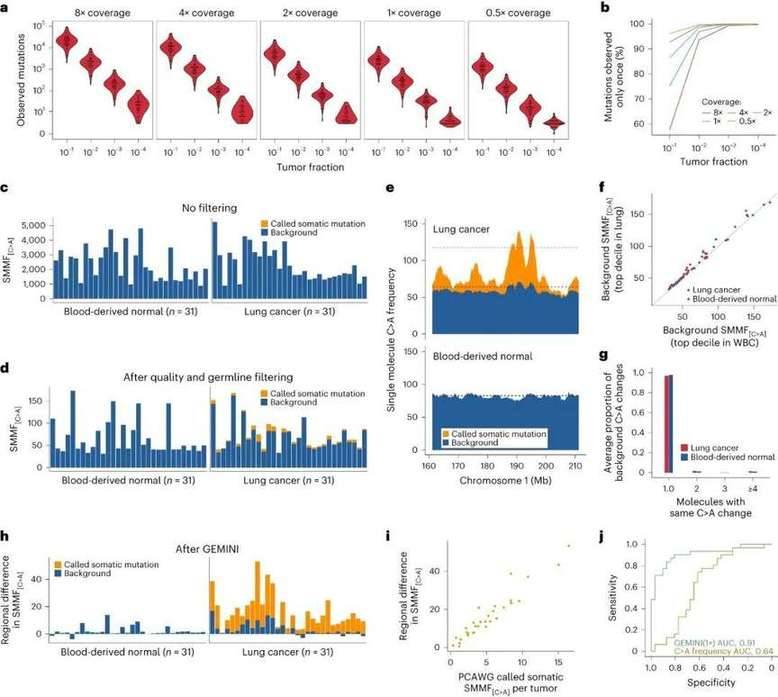 Single-molecule mutation analyses of PCAWG lung cancers and normal samples. (Bruhm et al., 2023)
Single-molecule mutation analyses of PCAWG lung cancers and normal samples. (Bruhm et al., 2023)
Mutation Profiles Specific to Cancer Types in cfDNA
Subsequently, the group of researchers examined data from 365 subjects within the cohort, focusing on low-coverage plasma whole genome sequencing (WGS). The outcomes revealed significant similarity in the genomic regions displaying high mutation frequencies. These regions were predominantly situated within genomic areas linked to tissue-specific late replication periods in both tumor tissues and cfDNA extracted from patients afflicted with lung cancer, melanoma, and B-cell non-Hodgkin's lymphoma. Concurrently, mutation frequencies specific to particular tumor types and mutations were correlated with gene expression, genomic segmentation as gauged by methylation signature vector analysis, and the presence of the heterochromatin marker H3K9me3.
Subjects lacking cancer, or those without mutation types or regions displaying a notable enrichment for cancer-associated features, exhibited either a lack of these attributes or only exhibited weak correlations with them. Taken together, the aforementioned findings indicate a connection between the genome-wide mutation rate of cfDNA and chromatin organization. The GEMINI system can leverage this characteristic to identify circulating tumor DNA (ctDNA).
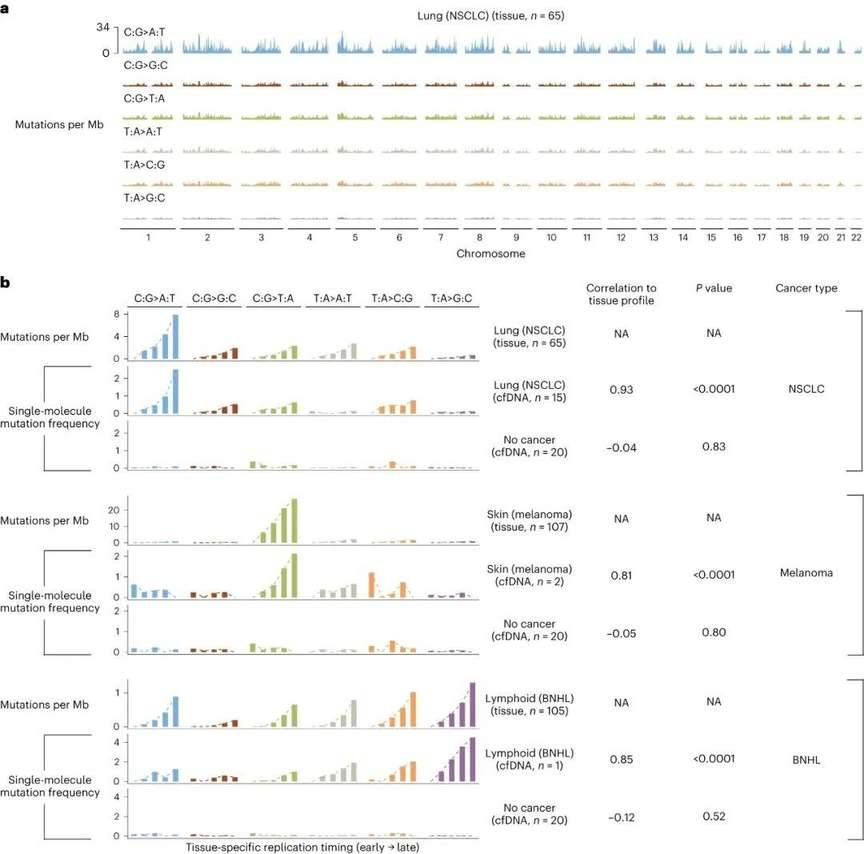 Genome-wide mutation profiles of tissue and plasma samples are associated with replication timing. (Bruhm et al., 2023)
Genome-wide mutation profiles of tissue and plasma samples are associated with replication timing. (Bruhm et al., 2023)
Summary
In essence, the team's discovery reveals that the utilization of single-molecule mutation profiles, acquired through cfDNA low-coverage WGS, holds promise for the non-invasive identification of cancer. Although the primary emphasis of this investigation was on identifying lung cancer within high-risk groups, it's worth noting that modified cfDNA mutation patterns were also detected in individuals with different cancers such as liver cancer, melanoma, and lymphoma. This implies that the approach might have a wider range of applications beyond its current scope.
Reference:
- Bruhm, Daniel C., et al. "Single-molecule genome-wide mutation profiles of cell-free DNA for non-invasive detection of cancer." Nature Genetics (2023): 1-10.


 Sample Submission Guidelines
Sample Submission Guidelines
