Nanopore Sequencing for Rapid Tumor Type Identification During Surgery
Introduction
Central nervous system (CNS) tumors, especially in children, rank among the most lethal forms of cancer. Surgical intervention typically serves as the primary treatment for this condition, posing a formidable challenge for neurosurgeons. Striking the right balance between maximizing tumor removal while minimizing neurological damage and surgery-related complications is paramount.
However, preoperative knowledge of the precise tumor type is often limited, and the current standard relies heavily on preoperative imaging and intraoperative frozen section histological analysis. Unfortunately, these methods occasionally provide inconclusive or incorrect diagnoses, necessitating pathologist intervention for a definitive diagnosis, typically a week post-surgery.
Beyond the issue of turnaround time, distinct neurological tumors demand tailored surgical resection strategies. For instance, complete resection yields limited prognostic improvement compared to near-total resection in the case of medulloblastoma, highlighting that maximal resection isn't always the preferred approach for such tumors. Conversely, some other tumors may benefit from an aggressive resection protocol. This dilemma underscores the risks of either excessive removal of healthy brain tissue leading to neurological damage or incomplete tumor removal necessitating a second surgery, imposing a substantial burden on the patient and their family.
The level of aggressiveness in neurosurgery is hence contingent on achieving an accurate and dependable tumor diagnosis.
Sturgeon and Rapid Nanopore Sequencing for Intraoperative Tumor Diagnosis
Jeroen de Ridder and his team at UMC Medical Center in Utrecht, the Netherlands, have redirected their focus towards Nanopore sequencing. The advantages of nanopore sequencing, including its cost-effectiveness, real-time data analysis, and expedited sample preparation, hold immense promise for diagnosing tumor subtypes during surgical procedures. Consequently, these Nanopore sequencing-based technologies permit early tissue sample analysis, enabling surgeons to adapt their neurosurgical strategies in response to prompt molecular diagnostic feedback.
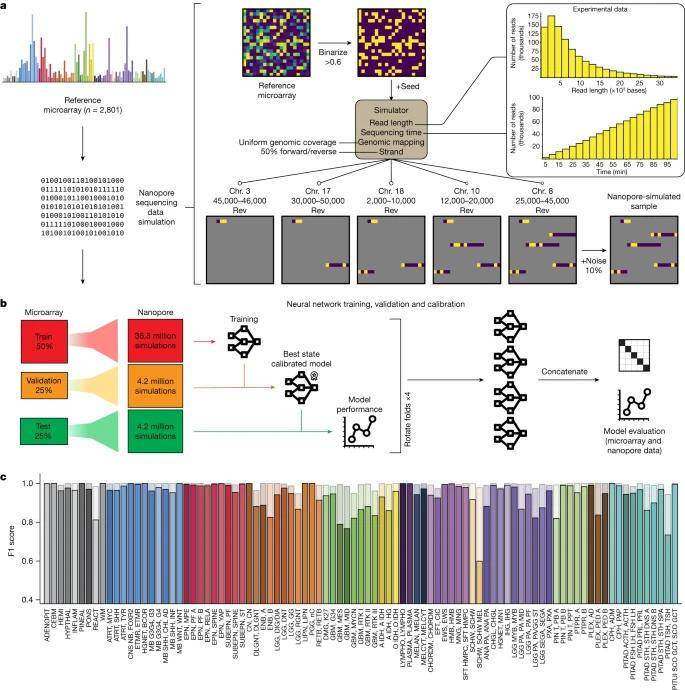 Schematic representation of the simulation, cross-validation approach and results on simulated data. (Vermeulen et al., 2023)
Schematic representation of the simulation, cross-validation approach and results on simulated data. (Vermeulen et al., 2023)
Yet, the practical challenge faced by Jeroen de Ridder and his team lies in the fact that neurosurgical decision-making often occurs within a narrow timeframe of less than 90 minutes. Sequencing within such constraints produces a somewhat 'sparse' methylation profile. Additionally, the identification of specific CpG sites in the genome that will be covered and generate methylation signals remains unpredictable.
To tackle these issues, the researchers have developed an advanced deep learning tool known as 'Sturgeon.' This patient-independent neural network classifier is finely tuned to handle sparse data, integrating cutting-edge technologies like machine learning, which is increasingly integral to the Nanopore sequencing data analysis toolkit.
What sets Sturgeon apart is its substantial allocation of computational resources for training and validating intricate neural networks before surgery. This distinguishes it from existing classification algorithms, which depend on intraoperative patient-specific model training. Following training on 36.8 million simulated Nanopore runs and validation on an additional 4.2 million simulated nanopore sequencing runs, Sturgeon exhibits remarkable speed, taking mere seconds to run on a laptop and deliver a precise tumor type determination using Nanopore-generated methylation data.
The researchers tested Sturgeon's performance by applying it to a retrospective CNS tumor dataset comprising 50 CNS tumor specimens and 415 publicly available nanopore sequencing data. Impressively, the model accurately classified the majority of samples (45 out of 50) based on the equivalent of 20-40 minutes of sequencing data, correctly classifying 92.2% (383 out of 415) of the samples overall. This demonstrates the model's consistent and reliable performance.
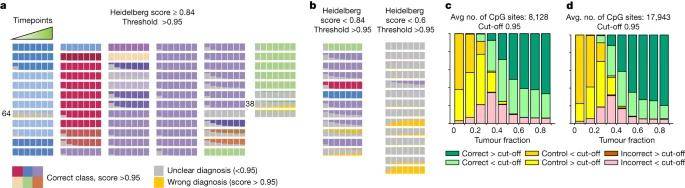 Classification performance over time on nanopore runs simulated from paediatric CNS tumour methylation arrays. (Vermeulen et al., 2023)
Classification performance over time on nanopore runs simulated from paediatric CNS tumour methylation arrays. (Vermeulen et al., 2023)
To further confirm the practicality of Sturgeon in conjunction with Rapid Nanopore Sequencing, the researchers implemented and evaluated the entire workflow in a real-world setting, involving 25 CNS tumor resections. They successfully completed the entire process, from specimen collection to tumor classification and diagnosis, within a 90-minute timeframe, achieving 18 correct intraoperative diagnoses—an ideal timeframe for guiding surgical decisions.
Nanopore Sequencing Technology
Nanopore sequencing stands at the forefront of contemporary gene sequencing technologies, featuring remarkable attributes, including real-time sequencing, user-friendly operation, and swift throughput. Its applications extend beyond whole genome sequencing and transcriptome research, notably excelling in the direct analysis of methylation. One of its inherent advantages lies in obviating the necessity for nucleic acid PCR, enabling seamless methylation analysis without additional sample preparation. Additionally, it unveils methylation insights in genomic regions that may elude or challenge traditional sequencing methods.
Reference:
- Vermeulen, C., et al. "Ultra-fast deep-learned CNS tumour classification during surgery." Nature (2023): 1-8.


 Sample Submission Guidelines
Sample Submission Guidelines
