Exome Sequencing Reveals the Impact of Rare Coding Gene Mutations on Adult Cognitive Function
Cognitive function is a complex mental process that includes attention, memory, processing speed, spatial ability, language and problem-solving skills and is difficult to assess in isolation. It has been shown that adult cognitive function is strongly influenced by genetics. Genome-wide association studies (GWAS) based on common variants have now identified nearly 4,000 genetic loci for cognitive function with small individual effects. Also, GWAS has demonstrated a shared genetic role between cognitive function and neurodevelopmental disorders, and large-scale exome studies have identified hundreds of potential genes. However, apart from the deleterious effects of exome-wide rare protein truncation variant (PTV) loading, no studies have systematically examined the effects of rare coding variants on cognitive phenotypes in adult populations.
Exome Sequencing Reveals Gene Mutations on Adult Cognitive Function
An article titled "The impact of rare protein-coding genetic variation on adult cognitive function" was recently published in Nature Genetics. To further explore more genes associated with cognitive phenotypes and to better understand the shared genes between cognitive function and neurodevelopmental disorders in adults, the team analyzed exome sequencing and genome-wide genotyping data from 454,787 participants in the UK Biobank (UKB) and examined cognitive function. The results showed that cognitive function in adults is significantly affected by mutations in rare protein-coding genes and identified eight genes (ADGRB2, KDM5B, GIGYF1, ANKRD12, SLC8A1, RC3H2, CACNA1A and BCAS3) associated with the adult cognitive phenotype. In addition, the rare genetic architecture of cognitive function partially overlaps with that of neurodevelopmental disorders. The researchers analyzed the relevance of rare coding variants for cognitive function and revealed the distribution of single genes with high impact on cognitive function in the normal adult population.
The researchers annotated exome sequencing data for PTV, missense mutations and synonymous mutations from 454,787 UKB participants and identified rare coding variants with minor allele frequencies (MAF) < 10-5 following previous exome studies for cognition-related traits. Also, the researchers further annotated all variants based on gene intolerance to loss-of-function (LoF) and missense mutations, analyzing a total of 649,321 protein truncation variants, 543,793 missense mutations and 30,603,887 synonymous rare mutations.
The analysis found that exome-wide PTV and missense mutational load had a significant deleterious effect on cognitive function, as reflected by lower EDU, longer RT and lower VNR scores. By analyzing 397,434 EUR samples in UKB, researchers identified eight genes associated with one or more significant cognitive phenotypes: KDM5B, ADGRB2, GIGYF1, SLC8A1, BCAS3, ANKRD12, RC3H2, and CACNA1A. consistent with expectations, PTV in eight genes showed deleterious effects.
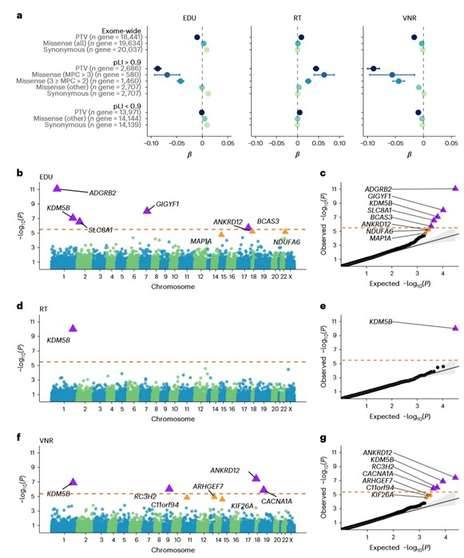 Impact of exome-wide burden of rare protein-coding variants and gene discovery based on the PTV burden for EDU, RT and VNR in EUR samples in the UKB. (Chen et al., 2023)
Impact of exome-wide burden of rare protein-coding variants and gene discovery based on the PTV burden for EDU, RT and VNR in EUR samples in the UKB. (Chen et al., 2023)
To systematically assess whether the LoF of eight cognitive genes affect phenotypes other than cognitive function, researchers conducted a phenome-wide association study (PheWAS) of 3,150 phenotypes in the unrelated UKB-EUR sample and found that six of the eight cognitive function genes were pleiotropic. For example, the rare PTV in KDM5B was not only strongly associated with three cognitive function phenotypes, but was also shown to be associated with muscle function, skeletal phenotypes, and bipolar disorder (BD).
Researchers performed PTV analysis on 13,011 gene sets in the UKB molecular signature database and identified 182, 66, and 56 significant gene sets for EDU, RT, and VNR, respectively, which overlap highly with features of cognitive function GWAS, suggesting that rare and common mutations regulate cognitive function through similar mechanisms. In addition, the researchers explored the relationship between rare coding variants and polygenic cognitive function risk based on common variants. The analysis showed that the effects of PRS and rare coding variants are synergistic, with rare coding variants affecting EDU and VNR in addition to PRS.
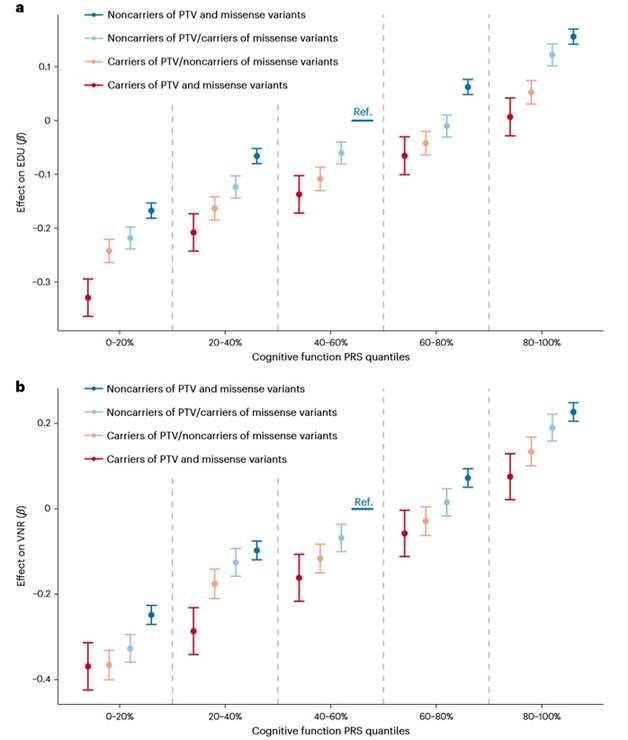
Researchers conducted a large-scale exome sequencing analysis of cognitive function phenotypes in the general adult population and found that increased exome-wide rare PTV was associated with lower cognitive function and identified eight genes associated with cognitive function in adults. The results of this study suggest that a subset of adults in the normal population have lower cognitive performance, which is caused by single gene defects. A particular advantage of exome studies is that genes and mutations identified by rare mutation testing tend to exhibit more significant effects than common mutations identified at GWAS. This study provides a new starting point for analyzing cognitive function and rare gene variants in populations, and a better understanding of how the genes and mutations reported in this study affect cognitive function and the biological basis of related diseases is still needed in the future.
Reference:
- Chen, Chia-Yen, et al. "The impact of rare protein coding genetic variation on adult cognitive function." Nature Genetics (2023): 1-12.


 Sample Submission Guidelines
Sample Submission Guidelines
