Bisulfite Genomic Sequencing (BSP-Seq) - Methylation Identification for ssDNA and dsDNA
One of the major mechanisms of epigenetic modification is DNA methylation. It has a substantial influence on gene expression which directly affects cellular activity. It is an epigenetic event in modulating genomic imprinting, embryonic development, cellular differentiation, X inactivation, and cell proliferation. However, abnormal DNA methylation is commonly associated with DNA and genomic instability leading to the development of diseases such as cancer. When DNA methylation occurs at certain vital cancer-related genes, it could lead to tumor suppressor gene silencing and tumorigenesis. This creates a crucial demand for highly sensitive and reliable methods to explore innovative diagnostic and therapeutic strategies. Illuminating the regions of DNA methylation facilitates the investigation of cell development, transcriptional silencing, and biomarker discovery.
Bisulfite genomic sequencing utilizes sodium bisulfite for DNA conversion which revolutionizes DNA methylation analysis. However, treatment with bisulfite is a harsh process that converts a stable double-stranded DNA into fragmented single strands with most cytosines converted to uracil. These conversions pose concerns and adjustments on UV spectrophotometry, PCR, and agarose gel electrophoresis.
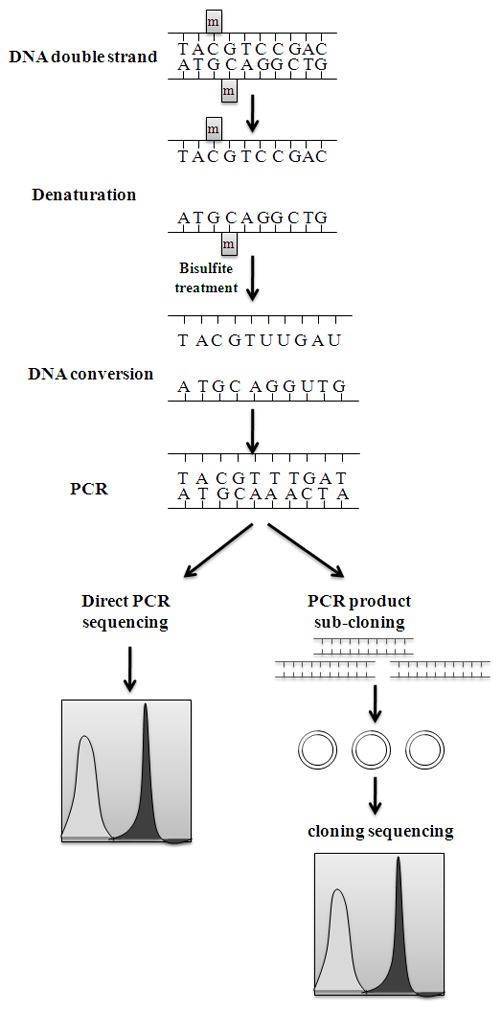
Figure 1. Bisulfite conversion and sequencing of samples (Li, 2011)
Methylation on a DNA strand could be detected when unmethylated cytosines in single-stranded DNA are converted into uracil residues which are recognized as thymine in subsequent PCR amplification. On the other hand, methylated cytosines are immune to this conversion and remain as cytosines allowing methylated Cs to be distinguished from unmethylated ones. Determining the methylation status in the loci of interest requires a subsequent PCR reaction using specific methylation primers after the treatment of sodium bisulfite. The status of methylation can be known either through direct PCR product sequencing or sub-cloning sequencing. However, direct PCR product sequencing generally yields poor quality and can confound quantification; hence, cloning followed by sequencing is recommended. Additionally, pyrosequencing can be used as an alternative for direct PCR sequencing.
Bisulfite sequencing analysis can not only identify methylation along the DNA single-strand but also detect the patterns of methylation on DNA double strands since the amplification products can be measured individually and the converted DNA strands are no longer self-complementary. Figure 1 shows the highlights of the DNA conversion process. A UV spectrophotometer is used for the quantification as RNA of the converted DNA. For the assessment of quality, the converted DNA will be run on a 2% agarose gel electrophoresis. The bands will appear as smears from 1500 to 100 bp. Bisulfite PCR primers are longer while amplicons are shorter than normal. The Whole-Genome Bisulfite Sequencing (WGBS) and Reduced Representation Bisulfite Sequencing (RRBS) are the two approaches that could be used to assess DNA methylation using NGS.
Reference:
- Li, Y., Tollefsbol, T. O. DNA methylation detection: bisulfite genomic sequencing analysis. Methods in molecular biology, 2011, 791, 11–21.


 Sample Submission Guidelines
Sample Submission Guidelines
