What is Pan-genome?
Pan-genome refers to all genes of the same species, including the Core genome, which is present in all individuals, and the individual-specific dispensable genome.
With the rapid development of long-read sequencing, Hi-C technology and computer algorithms, the reference genomes of each species have been continuously reported, but these genomes are usually constructed based on a representative individual, and a single linear reference genome cannot represent all the genetic variation information of the whole species. Therefore, constructing pan-genomes has become a new reference for high-quality reference genomes.
If you want to know more information about our Pan-genome Service, please contact our technique team.
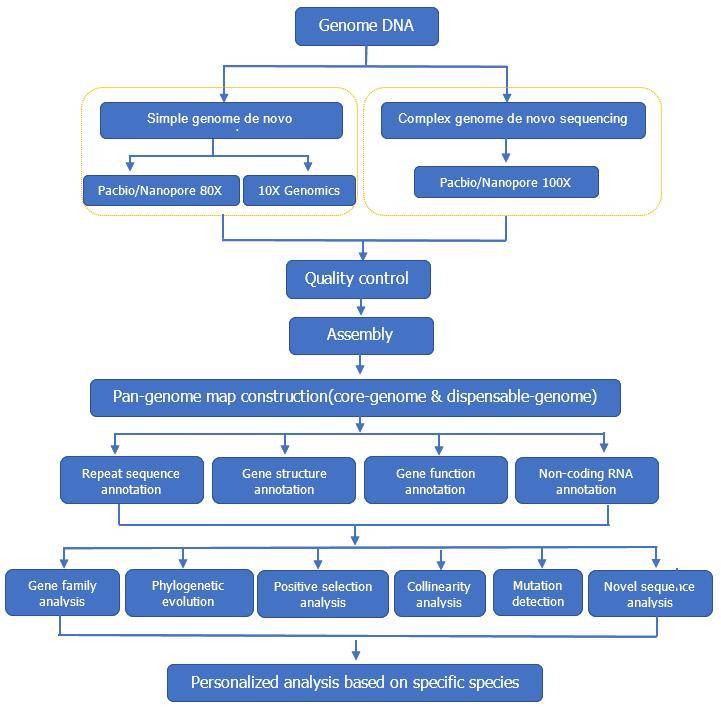 CD Genomics Pan-genome service workflow
CD Genomics Pan-genome service workflow
Or you may be interested in our article Pan-genome: Definition, Sequencing Methods, and Applications.
How to Build Pangenomes?
(1) Iterative Assembly
- In this approach, downstream data from multiple samples is meticulously compared with a reference genome.
- Unmatched reads are skillfully assembled into new contigs, seamlessly augmenting the original reference sequences.
- The amalgamation of these contigs forms the foundation of the pan-genome for the species.
(2) De Novo Assembly
Individual genomes are subject to de novo assembly and annotation, meticulously scrutinizing structural variations (SV), single nucleotide polymorphisms (SNP), insertions and deletions (InDel), copy number variations (CNV), and presence-absence variations (PAV) at a comprehensive whole-genome scale.
(3) Graph-Based Assembly
- The graph-based pan-genome methodology extends from genome de novo assembly and employs directed graphs to partition the species genome into the core and variable components.
- Contrasting with iterative assembly and genome de novo approaches, the graph-based pangenome approach integrates variation information from multiple genomes.
- This integration results in a more comprehensive representation of the species' genetic information, facilitating precise exploration of genetic variations.
- Noteworthy is the recent rapid development in research on graphical pangenomes, with publications emerging for various species such as cotton, tomato, lycopodium, cucumber, and others.
How to Design the Pan-genome Research Project?
Constructing a pan-genomic map not only enriches our understanding of a species' genetic makeup but also enables the characterization of individual or population variations by comparing sequenced individuals with the reference genome. This spans from simple single nucleotide polymorphisms (SNPs) and insertion-deletion mutations (InDel) to more complex structural variations (SV), copy number variations (CNV), and presence/absence variations (PAV). Furthermore, a comparative analysis of the functions and properties of core/non-core genes provides insights into the shared and unique phenotypic traits of species.
- Plant Pan-Genomic Research
In the realm of plants, higher plants exhibit substantial intraspecific genetic diversity to adapt to diverse growth environments. Leveraging constructed species pangenomes, we can reconstruct phylogenetic relationships between cultivated and wild species, exploring recombination and cascade effects at the population level. Using the distribution of variants in the pangenome, combined with transcriptomic analysis, we can detect variant expression and identify trait-related variants, correlating them with phenotypic information. Pan-genomic research-derived variant information can further fuel GWAS analysis, unveiling loci associated with agronomic traits grounded in structural variations, thus contributing valuable genetic resources for molecular breeding.
Please refer to our article A Review of Crop Pangenome for more information.
- Animal Pan-Genomic Research
In comparison to plants, animal pan-genome research is currently more limited, with a primary focus on humans and domesticated animals. By constructing large-scale population genomes and concentrating on species sharing common features, we can delve into differences in structural features, evolutionary history, convergent evolution, group characteristics, genome evolution, and potential functions. Developing pan-genomes for various strains, subspecies, or varieties allows for comparisons of genotypic differences across populations in different geographical regions, unveiling insights into species' environmental adaptations.
Please refer to our article A Review of Animal Pangenome for more information.


 Sample Submission Guidelines
Sample Submission Guidelines
