Metagenomic Sequencing in Phosphorus Cycling Genes Identification
Introduction
Phosphorus (P), an elemental cornerstone of life, plays an indispensable role in the functioning of all living organisms. This vital element is a fundamental component of DNA, RNA, ATP (adenosine triphosphate), and various other essential biomolecules. In plants, phosphorus is absorbed in the form of phosphates and then seamlessly integrated into organic compounds. In animals, it assumes a crucial role in the structure of bones, teeth, and a myriad of biological processes. However, the availability of phosphorus in terrestrial ecosystems has steadily declined over millennia due to its gradual loss through runoff. Soil microbial biomass studies have unequivocally demonstrated that diminished phosphorus concentrations in soil hinder both plant growth and the proliferation of essential soil microorganisms. Phosphorus transformations can be categorized into short-term processes driven by chemical, biological, or microbial mechanisms. Nevertheless, in the grand tapestry of the long-term global phosphorus cycle, the predominant transfers are dictated by the relentless forces of tectonic movements spanning geological epochs.
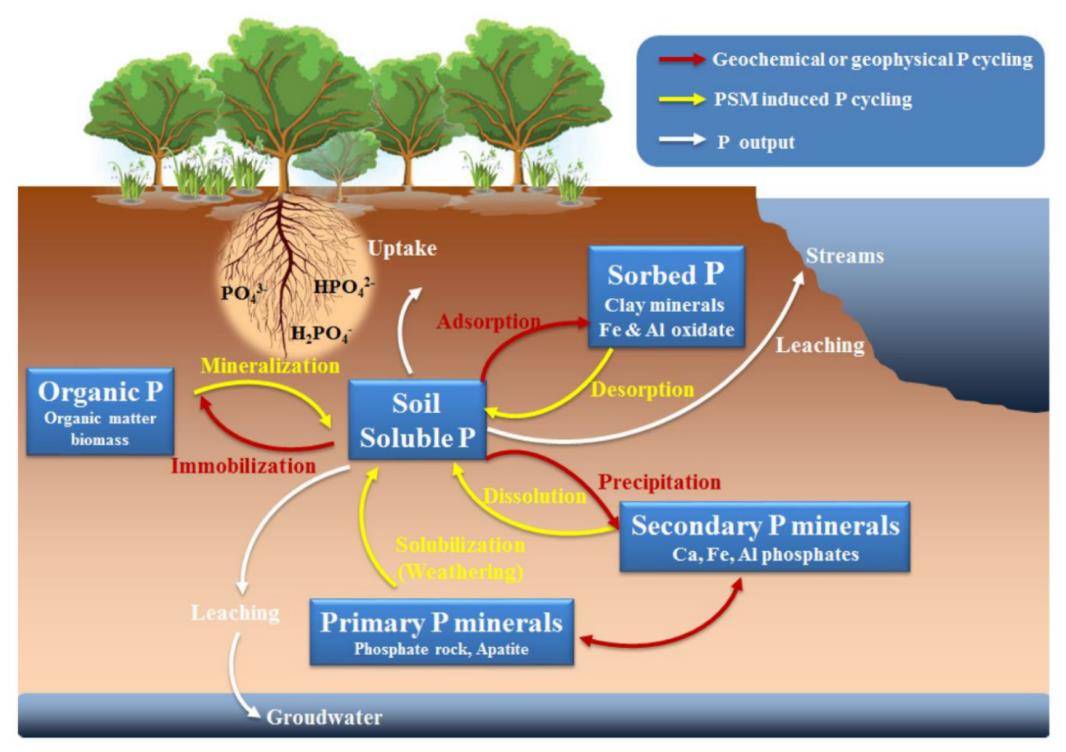 A schematic diagram of soil phosphorus (P) biogeochemical cycles. (Tian et al., 2021)
A schematic diagram of soil phosphorus (P) biogeochemical cycles. (Tian et al., 2021)
Metagenomic Sequencing Methodologies
Metagenomic sequencing involves the systematic analysis of DNA extracted from environmental samples, such as soil, sediment, water, or even the human gut, without the need for cultivation or isolation of individual organisms. This non-culture-based approach captures the genetic diversity of entire microbial ecosystems, shedding light on the collective genomic potential of these communities.
Key Advantages of Metagenomic Sequencing in Phosphorus Cycling Studies
- Unbiased Exploration: Unlike targeted gene sequencing, metagenomics provides an unbiased view of all genetic material present in a sample. This means that researchers can explore not only known phosphorus cycling genes but also discover novel genes and pathways that may play crucial roles in phosphorus metabolism.
- Environmental Relevance: Metagenomic sequencing allows scientists to examine genes within the context of their natural environment. This is particularly important for understanding how phosphorus cycling genes function in complex ecosystems, where interactions between different organisms can have profound effects on nutrient dynamics.
- High Throughput: Modern metagenomic sequencing technologies offer high-throughput capabilities, enabling the analysis of vast amounts of genetic data in a relatively short time. This scalability is essential for comprehensively characterizing the genetic diversity of complex microbial communities.
- Functional Potential Assessment: Metagenomics not only identifies genes but also provides insights into their functional potential. By annotating genes with known functions and metabolic pathways, researchers can infer the roles of specific genes in phosphorus cycling processes.
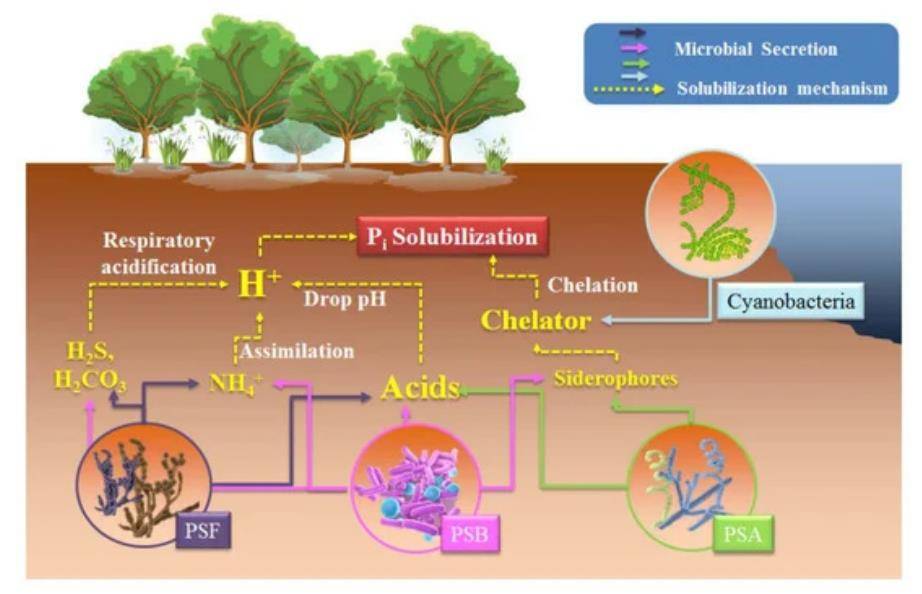 A schematic diagram of possible inorganic P (Pi) solubilization mechanisms in phosphate solubilizing microorganisms (PSM). (Tian et al., 2021)
A schematic diagram of possible inorganic P (Pi) solubilization mechanisms in phosphate solubilizing microorganisms (PSM). (Tian et al., 2021)
Unearthing Phosphorus Cycling Genes from Metagenomics
The quest to unravel the enigmatic world of phosphorus cycling involves a multifaceted approach, encompassing the following interrelated tasks:
1. Identification of Phosphorus Cycle-Related Genes: At the core of our endeavor lies the crucial task of identifying genes intricately entwined in the phosphorus cycle. These genes serve as the molecular architects orchestrating the complex dance of phosphorus through ecosystems.
2. Quantification of Related Gene Abundance: Understanding the quantitative aspects of phosphorus cycle-related genes is paramount. Accurate quantification provides insights into the dynamics of phosphorus cycling within various ecological niches.
3. Species Distribution of Related Genes: Investigating the distribution of these pivotal genes across different species offers profound insights into the evolutionary and ecological dimensions of phosphorus cycling.
Exploring Phosphorus-Related Genes in the KEGG Database
For an even broader perspective, the KEGG database provides a vast repository of phosphorus-related information. Here, you can access:
Matching Amino Acid Sequences, Nucleic Acid Sequences, and HMM Profiles: With a robust collection of data encompassing 125 KO (KEGG Orthology) immediate homolog clusters organized by gene symbols, the KEGG database opens up a world of possibilities for researchers eager to delve deeper into the genomics of phosphorus cycling.
Case: Impact of Land Use and Microbial Taxonomy on Soil Phosphorus Cycling Genes:
Background
Phosphorus (P) is vital for soil fertility and productivity. Microbes play a key role in adapting to P scarcity by using various genes related to P cycling. They investigated 23 such genes in forest, grassland, and cropland soils through metagenomic sequencing, examining their redundancy within ecosystems and genomic potential across microbial taxa.
Results
- Land Use Effect: Cropland and grassland soils had higher potential for mineralizing organic P and solubilizing inorganic P compared to forests. Genes for alkaline phosphatases and gluconic acid production were more common in these environments.
- Phosphonate Cycling: Forest soils showed greater potential for mineralizing phosphonates using C–P lyases, distinguishing their role in P cycling.
- Taxonomic Differences: Proteobacteria and Euryarchaeota had the greatest genomic potential to respond to P scarcity, while fungi played a more limited role.
- Key Functions: Phosphatase production, P solubilization, and P starvation response regulation demonstrated high intermetagenome redundancy, highlighting their importance in P cycling.
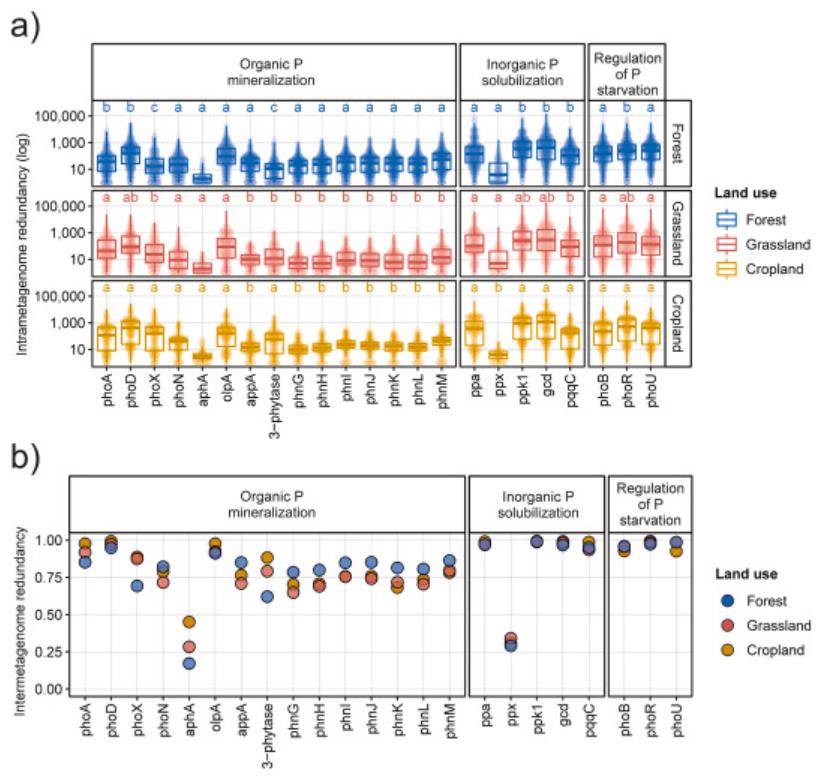 Intrametagenome redundancy as the number copies of P cycling genes. (Siles et al., 2022)
Intrametagenome redundancy as the number copies of P cycling genes. (Siles et al., 2022)
References:
- Siles, José A., et al. "Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining." Soil Biology and Biochemistry 174 (2022): 108826.
- Tian, Jiang, et al. "Roles of phosphate solubilizing microorganisms from managing soil phosphorus deficiency to mediating biogeochemical P cycle." Biology 10.2 (2021): 158.


 Sample Submission Guidelines
Sample Submission Guidelines
