How to Study DNA Hydroxymethylation (5hmC) in Plant Genomes?
Epigenetic modifications play a pivotal role in regulating gene expression and various biological processes in plants. Among these modifications, DNA hydroxymethylation (5-hydroxymethylcytosine or 5hmC) has garnered significant attention due to its emerging importance in plant development, stress responses, and environmental adaptation. Understanding the distribution and dynamics of 5hmC in plant genomes can provide crucial insights into epigenetic regulation and gene expression patterns. In this article, we will explore the methods and techniques used to study DNA hydroxymethylation in plant genomes.
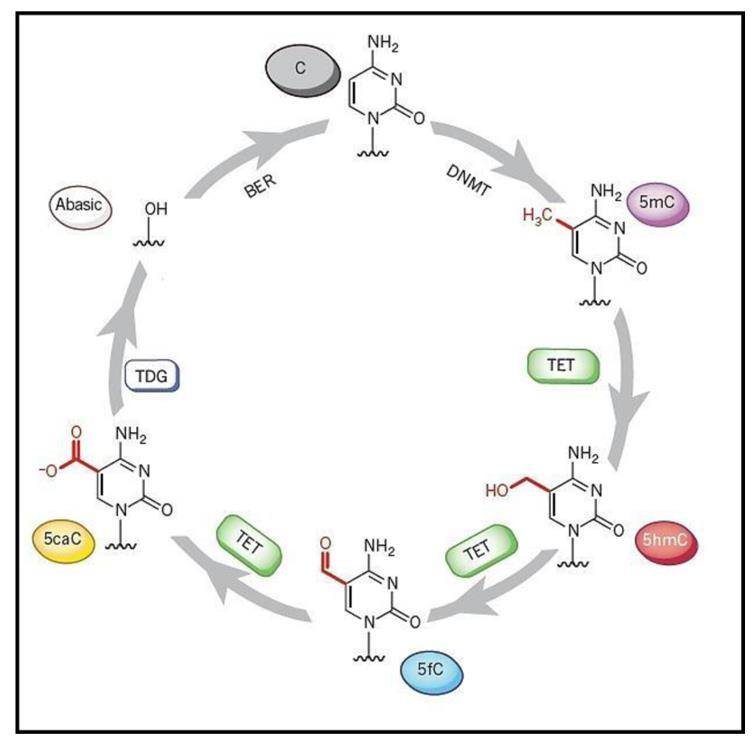 A putative cycling of cytidine derivatives via methylation and oxidative methylation. (Mahmood et al., 2019)
A putative cycling of cytidine derivatives via methylation and oxidative methylation. (Mahmood et al., 2019)
The Significance of DNA Hydroxymethylation (5hmC) in Plants
DNA hydroxymethylation is an epigenetic modification that results from the enzymatic oxidation of 5-methylcytosine (5mC) by the ten-eleven translocation (TET) family of enzymes. It has been identified as an intermediate step in the active demethylation process or as a stable epigenetic mark with distinct regulatory functions. The presence of 5hmC in plant genomes has been associated with key developmental processes, stress responses, and the regulation of transposable elements. As such, understanding the distribution and functional roles of 5hmC is crucial for unraveling the complexities of plant biology.
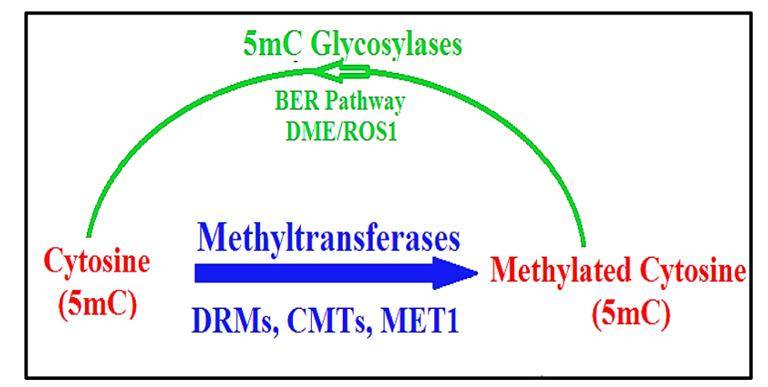 Putative enzymatic DNA demethylation pathway in plants. (Mahmood et al., 2019)
Putative enzymatic DNA demethylation pathway in plants. (Mahmood et al., 2019)
Immunoprecipitation-based Techniques
a. Utilization of 5hmC-Specific Antibodies: Immunoprecipitation, combined with highly specific antibodies targeting 5-hydroxymethylcytosine (5hmC), represents a robust approach for conducting comprehensive epigenetic analyses in plant genomes. This powerful method enables genome-wide profiling of 5hmC, elucidating its distribution and potential regulatory roles. Subsequent downstream analyses, such as quantitative PCR (qPCR) or high-throughput sequencing (ChIP-seq), facilitate precise mapping of genomic regions enriched with 5hmC, unraveling its associations with specific genetic elements, genes, or regulatory regions. Researchers investigating the epigenetic regulation of a specific plant developmental pathway use 5hmC-specific antibodies in immunoprecipitation to identify genomic regions where 5hmC is enriched during various developmental stages. Through subsequent ChIP-seq analysis, they discovered that 5hmC is significantly enriched in the regulatory regions of key genes involved in the pathway, suggesting its potential role in modulating gene expression during plant development.
b. 5fC and 5caC Immunoprecipitation (hMeDIP-seq): Building upon the success of 5hmC immunoprecipitation techniques, hMeDIP-seq involves the use of antibodies targeting 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) to capture these DNA modifications. By integrating immunoprecipitation with high-throughput sequencing, researchers gain valuable insights into the distribution and functional significance of 5fC and 5caC in the plant genome. A group of researchers studying the response of plants to environmental stressors uses hMeDIP-seq to investigate changes in 5fC and 5caC modifications under different stress conditions. They identify specific genomic loci where 5fC and 5caC levels change upon exposure to stress, potentially implicating these modifications in the plant's adaptive response to environmental challenges.
Chemical Labeling and Capture
a. Chemical Labeling and Affinity Purification (hMe-Seal): The hMe-Seal method employs chemical labeling of 5hmC using biotinylated glucosides, followed by affinity purification with streptavidin beads. This selective enrichment of 5hmC regions enables researchers to gain a deeper understanding of its distribution and functional implications within the plant genome. By harnessing sequencing-based approaches, hMe-Seal allows for detailed characterization of 5hmC-enriched regions, thus unraveling its role in gene regulation and genome stability.
b. Oxidative Bisulfite Sequencing (oxBS-Seq): This novel technique capitalizes on the selective chemical oxidation of 5hmC to 5-formylcytosine (5fC), followed by bisulfite treatment. Subsequent high-throughput sequencing allows for the discrimination between 5mC and 5hmC, providing a comprehensive view of the plant genome's hydroxymethylation landscape. oxBS-Seq offers valuable insights into the interplay between DNA methylation and hydroxymethylation, shedding light on their combinatorial effects on gene expression and plant development.
Single-Molecule, Real-Time (SMRT) Sequencing
SMRT Sequencing with Direct Detection of Modified Bases: Pioneered by PacBio, SMRT sequencing stands at the forefront of long-read sequencing technologies. This cutting-edge approach enables the direct detection of modified bases, including 5hmC, without the need for bisulfite treatment. With its extended read lengths, SMRT sequencing excels at studying complex repetitive regions, structural variations, and epigenetic modifications within the plant genome. This empowers researchers to unravel the intricate epigenetic landscapes, identify 5hmC-associated regulatory elements, and decipher the interplay between epigenetic modifications and plant phenotype.
Reference:
- Mahmood, Asaad M., and Jim M. Dunwell. "Evidence for novel epigenetic marks within plants." AIMS genetics 6.04 (2019): 070-087.


 Sample Submission Guidelines
Sample Submission Guidelines
