DNA Methylation Valleys (DMVs) in the Rice Genome: Epigenetic Regulation and 3D Genome Architecture
DNA methylation modification represents an evolutionarily conserved epigenetic process that plays a pivotal role in a diverse array of biological processes. One intriguing facet of this modification is the presence of DNA methylation valleys (DMVs), which denote continuous regions within the genome devoid of DNA methylation modification. Emerging evidence strongly suggests that DMVs wield significant influence over the regulation of biological growth and developmental processes.
Surprisingly, the exploration of DMVs within the rice genome has remained largely uncharted territory, leaving a significant gap in our understanding. Furthermore, the impact of DNA methylation on the intricate higher-order structure of rice chromatin remains an enigma.
In this study, they embarked on a comprehensive genome-wide characterization of DMVs within the rice genome. Employing a multi-faceted approach, they leveraged epigenomic data and three-dimensional genomic information obtained from both wild-type and osdrm2 mutant rice plants. The investigation delves into the intricate interplay between DNA methylation modifications and the higher-order architecture of rice chromatin.
In this comprehensive research endeavor, the authors embarked on a journey to meticulously dissect the landscape of DMVs within the rice genome. Their quest led them to a striking discovery: the presence of grand DMVs, substantial segments of genomic DNA that appear to originate from the integration of genome material into organelles. Remarkably, these grand DMVs exhibited an intriguing enrichment of genes closely linked to organelle functionality.
Conversely, the authors also unveiled another intriguing facet of DMVs: the existence of short DMVs (sDMVs). These compact DNA regions demonstrated a unique enrichment pattern, as they were found to harbor genes intricately associated with processes such as morphogenesis, transcriptional regulation, and RNA synthesis. What's even more fascinating is that these genes exhibited a tissue-specific expression pattern, adding an additional layer of complexity to their functional roles within the organism.
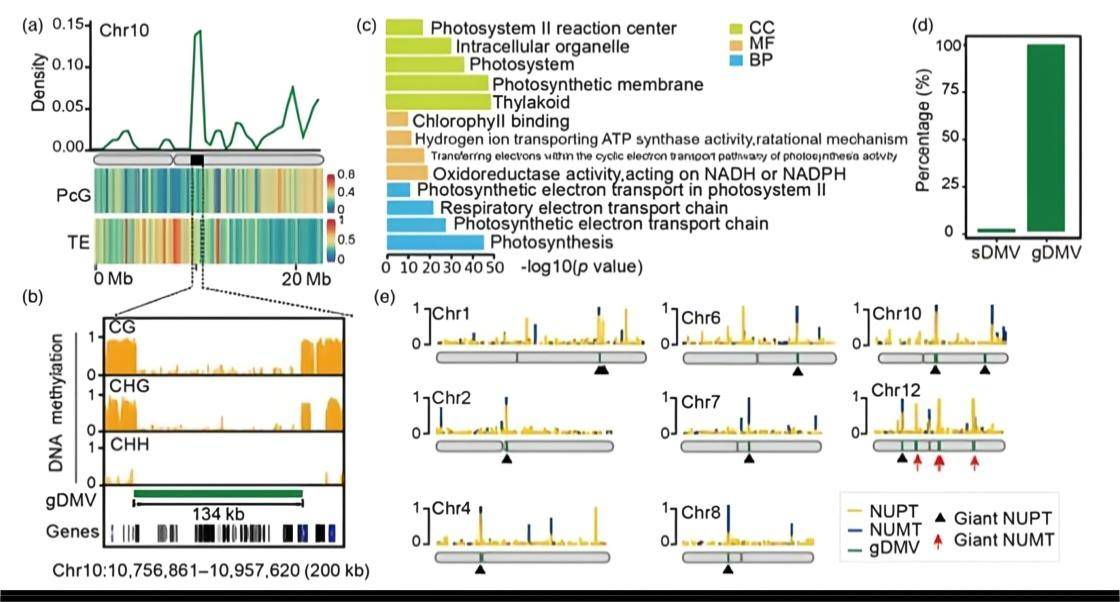 Uneven distribution of DMVs and gDMVs derived from NORG. (Zhang et al., 2023)
Uneven distribution of DMVs and gDMVs derived from NORG. (Zhang et al., 2023)
OsDRM2 plays a pivotal role in orchestrating non-CG methylation modifications within the rice genome, serving as the cornerstone for the establishment of DNA methylation through the RdDM pathway. By conducting a comprehensive comparison of genome-wide methylation profiles between the wild-type and osdrm2 mutant strains, the researchers have unveiled that OsDRM2 serves as a guardian of DNA methylation modifications along the sDMV boundary. The functional loss of OsDRM2 exerts a notable impact on the extent of the sDMV region. Additionally, a complementary analysis of eChIP-seq data pertaining to histone modifications in both wild-type and mutant strains has highlighted the enrichment of H3K27me3 and/or H3K4me3 modifications within the sDMV region. Notably, alterations in DNA methylation at the sDMV boundary in the osdrm2 mutant have subtle repercussions on the distribution of histone modifications within the confines of the sDMV region.
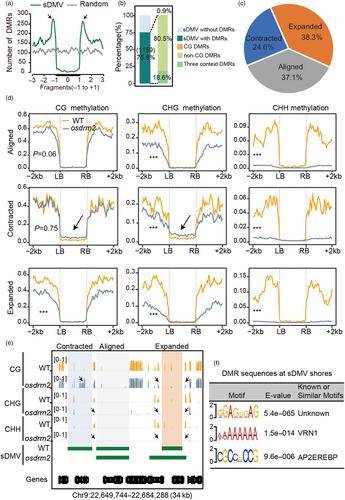 OsDRM2 maintains DNA methylation at the sDMV shores in rice. (Zhang et al., 2023)
OsDRM2 maintains DNA methylation at the sDMV shores in rice. (Zhang et al., 2023)
Furthermore, the researchers meticulously charted the 3D genome linked to H3K27me3 in both the wild type and osdrm2 mutant strains. Through a comprehensive analysis, they unveiled that the global A/B compartment structure remained unaltered at lower resolutions. However, at a finer scale, the chromatin exhibited a more relaxed configuration in the osdrm2 mutants, accompanied by a striking decline in long-distance chromatin interactions. Simultaneously, the diminished non-CG methylation levels at the sDMV boundary perturbed the intricate chromatin interaction network connected to the sDMV region.
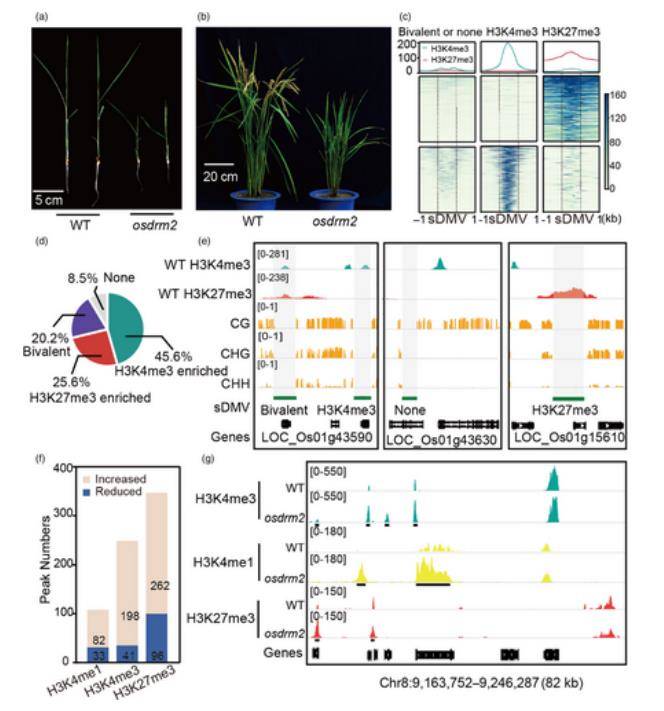 OsDRM2 ablation has a weak impact on histone modifications within sDMV regions. (Zhang et al., 2023)
OsDRM2 ablation has a weak impact on histone modifications within sDMV regions. (Zhang et al., 2023)
This study presents a systematic exploration of rice genomic differentially methylated regions (DMVs) while shedding light on the functional roles of DNA methylation modifications at the boundaries of these structural domains within the three-dimensional genomic landscape. These findings contribute to a more comprehensive understanding of the rice genome's structural intricacies, offer fresh perspectives into the impact of DNA methylation modifications on chromatin organization, and furnish a solid theoretical foundation for advancing genetic enhancements in rice.
Reference:
- Zhang, Wei, et al. "Domains Rearranged Methylase 2 maintains DNA methylation at large DNA hypomethylated shores and long-range chromatin interactions in rice." Plant Biotechnology Journal (2023).


 Sample Submission Guidelines
Sample Submission Guidelines
