Potato Pan-Genomics: Evolution and Hybrid Breeding Breakthroughs
With the advent of long-read sequencing technology, research concerning genomes has advanced to unprecedented heights. Not only has the completeness, continuity, and accuracy of genome assembly improved significantly in contrast to next-generation sequencing technology, but the breakthroughs span all aspects of genomics. This progress not only facilitates the in-depth investigation of individual high-quality genomes but also enables the pursuit of pan-genome research.
The potato stands as one of the globe's most vital non-cereal crops, and the vast majority of commercially cultivated potato strains display considerable tetraploid. The progression in diploid crossbreeding via authentic seeds holds the power to completely transform upcoming potato cultivation and breeding endeavors. Up until now, only limited investigations have delved into the genetic evolution and diversity of both wild and cultivated potatoes, constraining the practical utilization of potato genome diversity within breeding initiatives.
The researchers successfully compiled 44 meticulously detailed diploid potato genomes utilizing the PacBio HiFi and Hi-C technologies, culminating in a comprehensive pan-genomic exploration. This exploration is poised to expedite the advancement of hybrid potato breeding techniques and enhance our comprehension of potato's evolution and biological characteristics as a pivotal global food source.
Pan-Genome Exploration of Petota Varieties
This investigation encompassed the assembly of seven representative genomes at the chromosomal level, with a total of 24.5 Gb of HiFi reads obtained through high-throughput chromatin conformation capture (Hi-C) technology. The genome sizes of the assembled representatives ranged from 835.1 Mb to 1.71 Gb. Employing the Markov clustering algorithm, the anticipated 2,701,787 genes were clustered to create a pan-genome comprising 51,401 pan-genomic clusters.
Phylogenetic Analysis of Petota and Adjacent Species
To elucidate the evolutionary connections between Petota and its close relatives Lycopersicon and Etuberosum, PacBio long sequences were sequenced for the outgroups Solanum etuberosum and Solanum palustre. De novo assembly was executed, followed by the utilization of supermatrix and multispecies clustering techniques to deduce inter-species phylogenetic trees. Among the 1,899 evolutionary trees, 334 (17.6%) lent support to Etuberosum the sibling branch of Petota. Additionally, the D-test facilitated the identification of discernible gene exchange between Petota and Etuberosum.
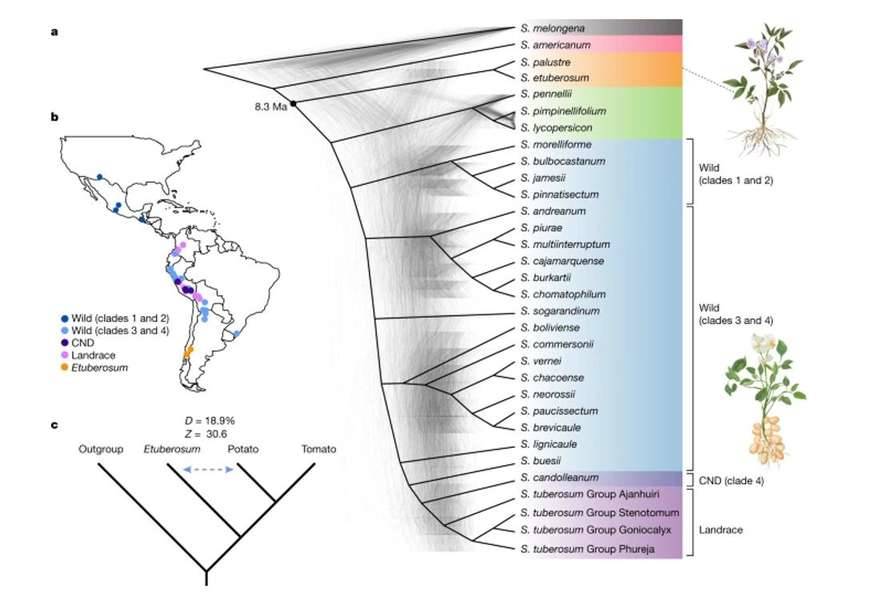 Geographical distribution and phylogeny of the Solanum genus. (Tang et al., 2022)
Geographical distribution and phylogeny of the Solanum genus. (Tang et al., 2022)
Broadening the Repertoire of Resistance Genes
Scientists devised a procedure for NLR annotation, which they validated using a tomato NLR dataset acquired through resistance gene enrichment sequencing. This effort yielded a substantial compilation of 57,683 NLR genes. The count of NLR copies displayed substantial diversity across different potato species. Predictions unveiled that Etuberosum and tomato genomes held between 280 and 344 NLRs, while the potato MTG assembly exhibited noticeable gene amplification.
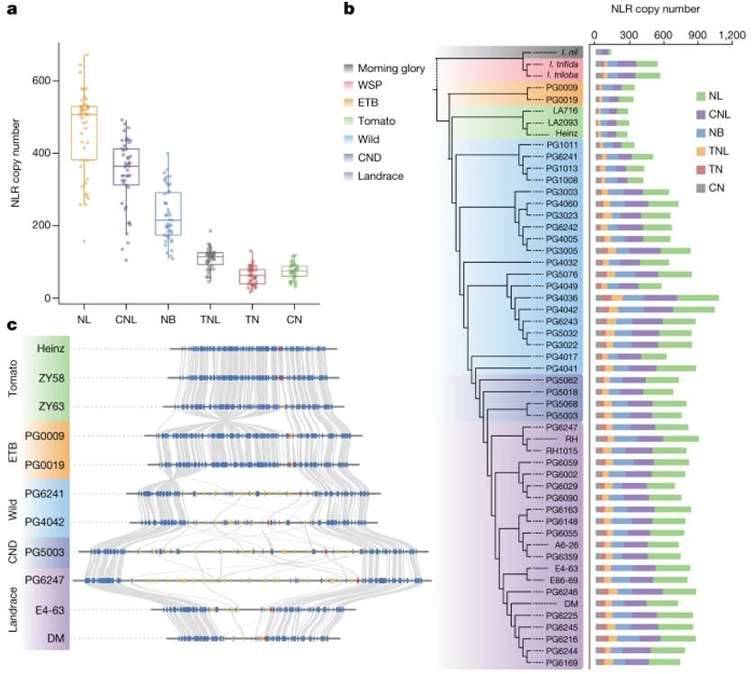 Evolution of resistance genes in potato. (Tang et al., 2022)
Evolution of resistance genes in potato. (Tang et al., 2022)
Tuber Homologous Genes
Regarding tuber homologous genes, this study uncovered a total of 149,663 CNSs specific to potatoes, spanning a length of 6.9 megabases. Among these, approximately 54.4% were situated within introns, suggesting a potential influence on the expression of 17,871 genes.
In the pursuit of identifying pivotal genes linked to tuber development, the present investigation pinpointed 732 genes with predominant expression in stolons or tubers. Of these, 229 genes were associated with potato-specific CNSs. Among these genes, 28 were identified as transcription factors, with a lone representation from the plant-specific TCP transcription factor family (Soltu.DM.06G025210). Transcriptome data revealed that this particular gene displayed prominent expression in potato stolons.
Pan-Genome-Guided Hybrid of Potato Breeding
Pioneering pan-genome-guided hybrid breeding for potatoes, the researchers unearthed a grand total of 561,433 high-confidence structural variations (SVs) exceeding 50 base pairs in size, of which roughly 55.5% were classified as rare. Through amalgamating data from 20 native species and 4 instances of S. candolleanum, an inversion map was meticulously constructed. An intra-arm inversion spanning about 5.8 megabases was mapped to chromosome 3. Within this inversion, a critical gene responsible for β-carotene hydroxylation, governing the accumulation of zeaxanthin and thus imparting the characteristic yellow hue to the tuber, was identified. This gene was intricately linked with 464 other genes residing within the inversion. Consequently, opting for individuals exhibiting yellow tuber flesh for nutritional attributes might inadvertently trigger pronounced resistance to unforeseen phenotypic traits. The established pan-genomic inversion map now empowers breeders with the capability to judiciously select appropriate donor or recipient strains for the process of backcrossing.
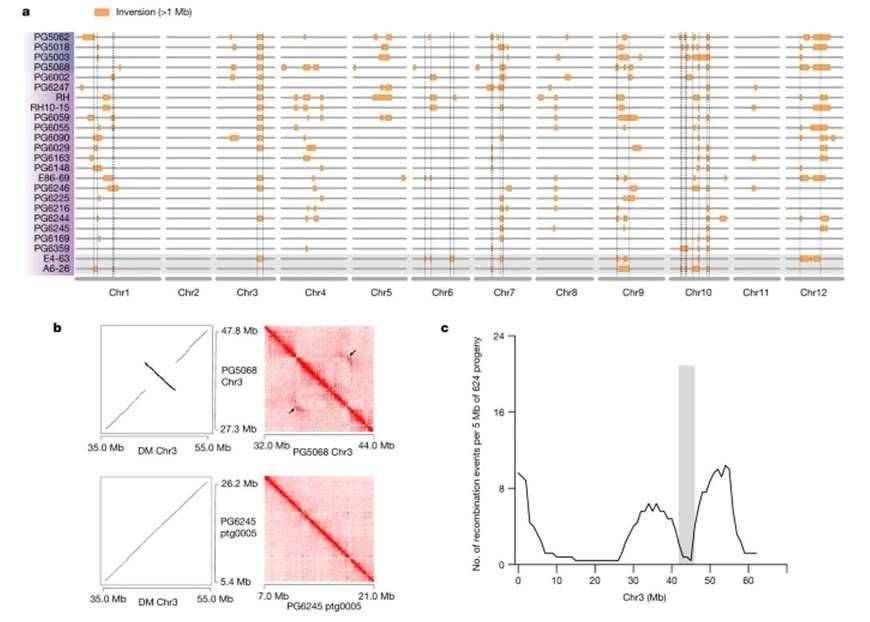 Pan-genome-based map of large inversions. (Tang et al., 2022)
Pan-genome-based map of large inversions. (Tang et al., 2022)
Summary
The study has unearthed 44 genomes of exceptional quality along with extensive genetic diversity. These findings serve as valuable assets for the comprehensive enhancement of potato breeding at a genome-wide level. Moreover, these resources hold immense potential for the creation of an encompassing pan-genomic reference that amalgamates genomes and variations sourced from 44 distinct potato germplasms. Notably, the revelation of IT1 and the intricate interplay involving its partner factor SP6A will play a pivotal role in establishing a solid groundwork for comprehending the evolutionary trajectory of tuber development.
As the PacBio platform undergoes expansion, the cost associated with HiFi sequencing experiences a reduction. This reduction in cost facilitates the incorporation of pan-genome construction into every project undertaken.
Reference:
- Tang, Dié, et al. "Genome evolution and diversity of wild and cultivated potatoes." Nature 606.7914 (2022): 535-541.


 Sample Submission Guidelines
Sample Submission Guidelines
