LncRNA Sequencing
CD Genomics is offering high-throughput and cost-efficient lncRNA sequencing service by combining the latest Illumina sequencing instruments and advanced bioinformatics analysis.
The Introduction of LncRNA Sequencing
Long non-coding RNAs (lncRNAs) are defined as a large and diverse class of transcribed RNAs with size greater than 200 nt that do not encode proteins. LncRNAs are widely distributed in organisms and lncRNA transcripts account for the major part of the non-coding transcriptome. LncRNAs may be classified into different subtypes (including antisenses, intergenic, overlapping, intronic, bidirectional, and processed) based on the position and direction of transcription in relation to other genes. LncRNAs resemble mRNAs because they are typically transcribed from active chromatin, polyadenylated, and capped. However, they do not direct protein synthesis.
LncRNAs are functionally important to organisms and not merely the product of transcriptional noise. A myriad of molecular functions of lncRNAs have been discovered in mammals and plants, including nucleosome repositioning, chromatin remodeling, transcriptional control, and posttranscriptional processing. LncRNAs are increasingly implicated in disease occurrence, genomic imprinting and developmental regulation. Gene expression profiling and in situ hybridization studies have revealed that lncRNA expression is developmentally regulated, can be tissue- and cell-type specific, and can vary spatially, temporally, or be in response to stimuli.
The application of next-generation sequencing technology has greatly facilitated the discovery and function analysis of lncRNAs. LncRNA sequence information can be acquired at single-base resolution via library preparation, high-throughput sequencing, and powerful bioinformatics analysis. We construct the sequencing library by the removal of rRNA and retain both lncRNAs and mRNAs. The lncRNA-mRNA interaction analysis contributes to the illumination of lncRNA regulatory networks.
Advantages of LncRNA Sequencing
- Identifies known and novel features
- Allows profiling of lncRNAs across a wide dynamic ranges
- Explores novel biomarkers and lncRNAs regulatory networks
Applications of LncRNA Sequencing
LncRNA sequencing can be used for but not limited to the following research:
- Discovering and Predicting Novel lncRNAs;
- Disease Investigation;
- Regulatory Network Analysis.
LncRNA Sequencing Workflow
The general workflow for lncRNA sequencing is outlined below. To construct lncRNA sequencing library, the first step of lncRNA sequencing is to deplete rRNA, followed by RNA fragmentation, cDNA synthesis, adaptor ligation, size selection and PCR enrichment. Our highly experienced expert team executes quality management, following every procedure to ensure confident and unbiased results.

Service Specification
Sample Requirements
|
|
Click |
Sequencing Strategies
|
| Data Analysis We provide multiple customized bioinformatics analyses:
|
Analysis Pipeline
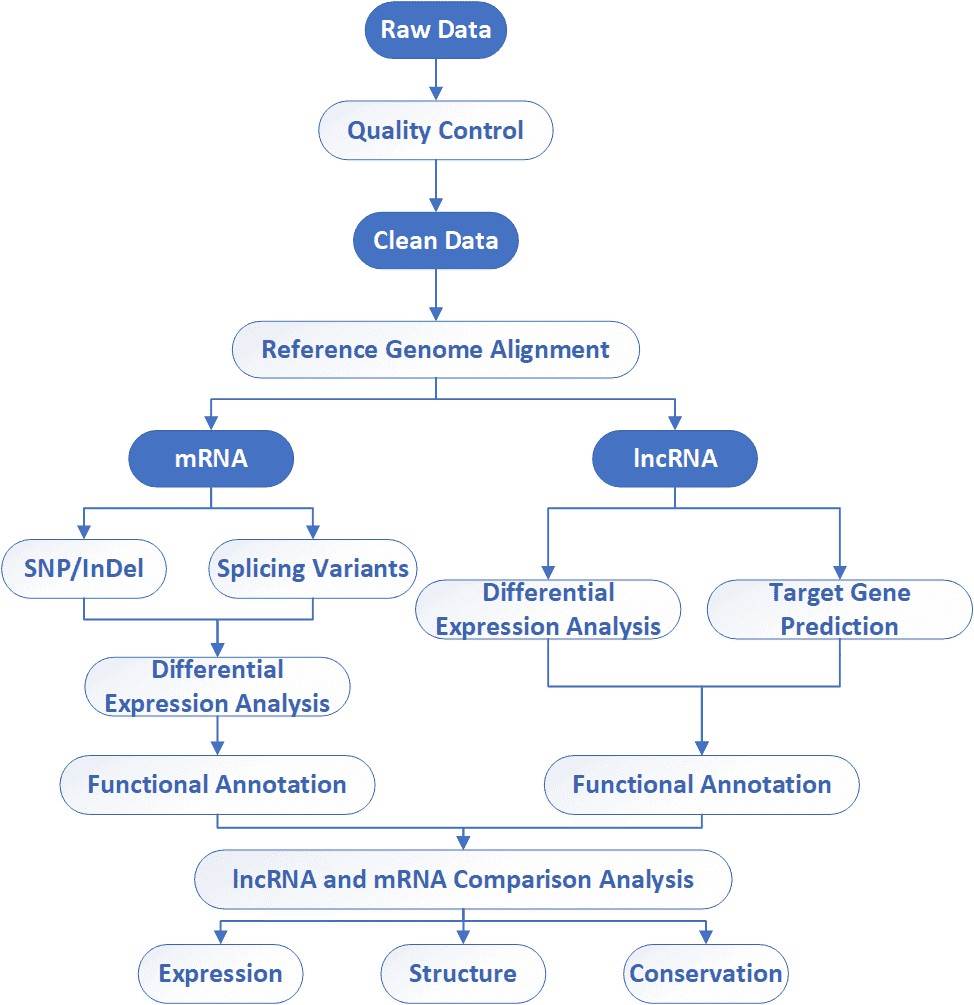
Deliverables
- The original sequencing data
- Experimental results
- Data analysis report
- Details in LncRNA Sequencing for your writing (customization)
Our PhD-level bioinformatics team provides comprehensive analysis for both lncRNAs and mRNAs, enabling access to lncRNA and mRNA information in a single sequencing run. We can help in the experimental design at the very beginning of your project and offer consultation at every stage of the project process.
Partial results are shown below:

Sequencing quality distribution

A/T/G/C Distribution

IGV Browser Interface

Correlation Analysis Between Samples

PCA Score Plot

Venn Diagram

Volcano Plot

Statistics Results of GO Annotation

KEGG Classification
1. What species are appropriate for lncRNA sequencing studies?
For lncRNA sequencing studies, the appropriate species need to meet the following requirements: (i) eukaryotes; (ii) at least scaffold-level reference genome available; (iii) relatively complete genome annotations.
2. Why remove rRNA when constructing lncRNA sequencing libraries?
Ribosomal RNA (rRNA) is the most highly abundant component of total RNA, comprising 80% to 90% of the molecules in a total RNA sample from animal or human. For efficient transcript detection, highly abundant rRNAs must be removed before sequencing.
3. How to predict lncRNA targets?
Both analyses of co-location and co-expression of protein-coding RNAs and lncRNAs have been proved effective in the investigations of the potential function of lncRNAs in biological processes and lncRNA target prediction. When the co-location analysis considers the adjacent coding genes maybe lncRNA targets, the co-expression analysis deems the co-expressed genes to be probable lncRNA targets, independent of location.
4. What are the differences between lncRNA and mRNA?
LncRNAs resemble mRNAs because they are typically transcribed from active chromatin, polyadenylated, and capped. However, they do not direct protein synthesis. Some differences between lncRNA and mRNA are summarized in the table below.
| mRNA | lncRNA |
| Protein coding transcript | Non-protein coding, regulatory transcript |
| Well conserved between species | Poorly conserved between species |
| Present in both nucleus and cytoplasm | Many predominantly nuclear, others nuclear and/or cytoplasmic |
| Total 20-24,000 mRNAs | Currently ~30,000 lncRNA transcripts, predicted 3-100 fold of mRNA in number |
| Expression level: low to high | Expression level: very low to moderate |
Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes
Journal: Molecular Metabolism
Impact factor: 6.799
Published: 23 August 2017
Abstract
The authors identified approximately 1500 novel lncRNAs, and some of them were differentially expressed in obese mice. Two lncRNAs (βlinc2 and βlinc3) are highly enriched in β-cells, correlated to body weight gain and glycemia levels in obese mice and modified in diabetic db/db mice. Moreover, the expression of the human orthologue of βlinc3 was changed in the islets of type 2 diabetic patients, associated to the BMI of the donors. Modulation of the level of the two lncRNAs by overexpression or downregulation in MIN6 and mouse islet cells increased β-cells but did not affect glucose regulation.
Materials & Methods
- Five-week old male C57BL/6 mice
- Mice islets
- RNA extraction
- Library construction
- RNA-sequencing
- Illumina HiSeq2000
- Differential expression analysis
- Differential analysis
- Gene ontology analysis
- Measurement of lncRNAs expression
Results
1. RNA sequencing analysis
RNA sequencing yielded 1558 novel lncRNAs, and some of them were differentially expressed in obese mice. Functional annotation showed enrichment for biological pathways related to protein localization and transport, redox processes, as well as intracellular transport.
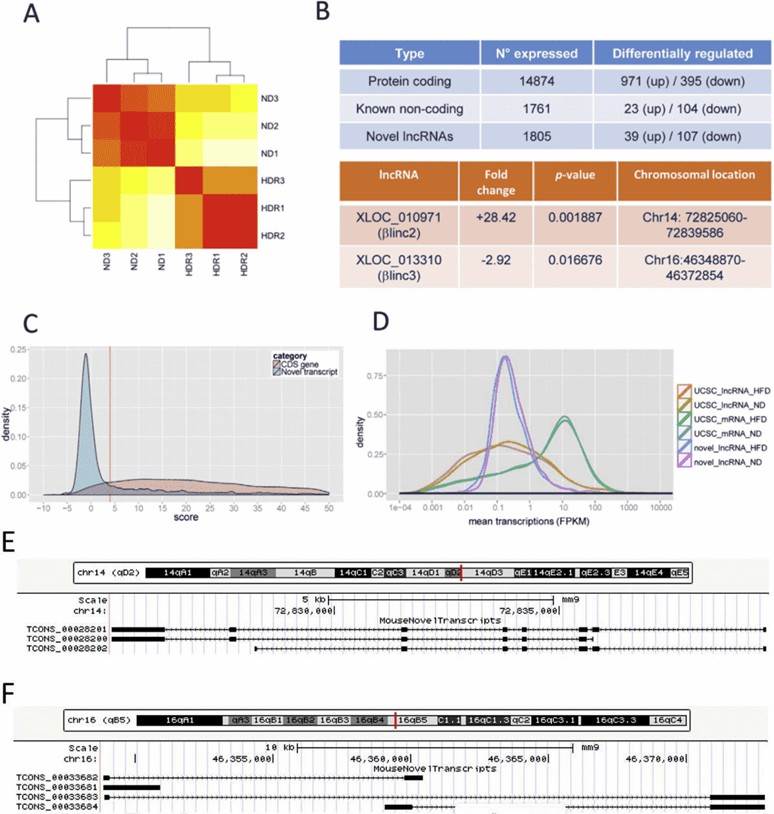 Figure 1. Overview of RNA-sequencing results. (A) Hierarchical clustering. Red represents no distance and yellow represents a longer distance. ND denotes normal diet, HDR denotes high-fat diet responders. (B) summary of differentially expressed genes. (C) coding potential for novel transcripts; (D) Size distribution of protein-coding genes, known and novel lncRNAs. (E & F) Locus architecture and isoforms of βlinc2 or βlinc3.
Figure 1. Overview of RNA-sequencing results. (A) Hierarchical clustering. Red represents no distance and yellow represents a longer distance. ND denotes normal diet, HDR denotes high-fat diet responders. (B) summary of differentially expressed genes. (C) coding potential for novel transcripts; (D) Size distribution of protein-coding genes, known and novel lncRNAs. (E & F) Locus architecture and isoforms of βlinc2 or βlinc3.
2. The expression levels of βlinc2 and βlinc3
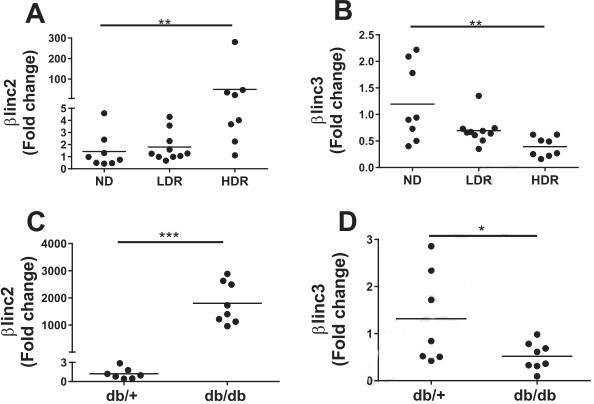 Figure 2. The expression levels of βlinc2 and βlinc3 are modified in islets from mice fed a high-fat diet and in db/db mice.
Figure 2. The expression levels of βlinc2 and βlinc3 are modified in islets from mice fed a high-fat diet and in db/db mice.
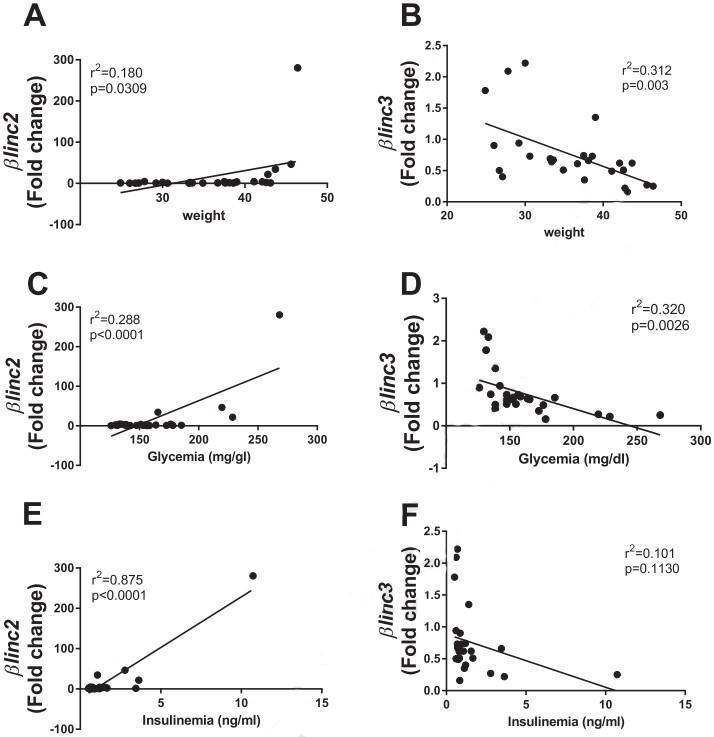 Figure 3. Correlations of the expression of βlinc2 and βlinc3 with body weight, and glycemia of C57BL/6mice.
Figure 3. Correlations of the expression of βlinc2 and βlinc3 with body weight, and glycemia of C57BL/6mice.
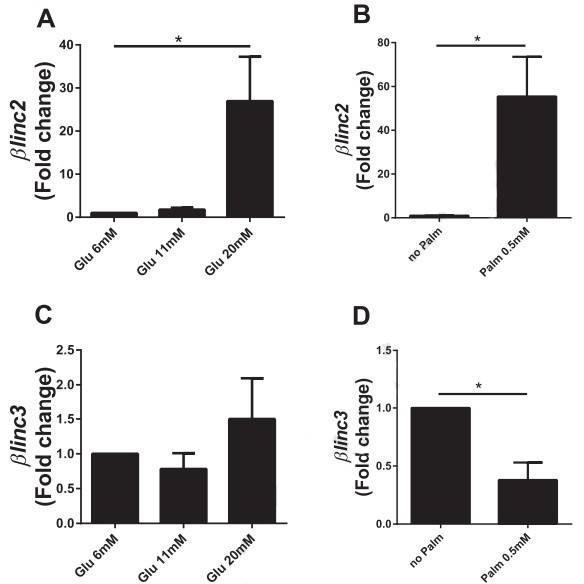 Figure 4. In vitro effects of chronically-elevated glucose and palmitate on the level of the two lncRNAs differentially expressed in islets from mice fed a high-fat diet.
Figure 4. In vitro effects of chronically-elevated glucose and palmitate on the level of the two lncRNAs differentially expressed in islets from mice fed a high-fat diet.
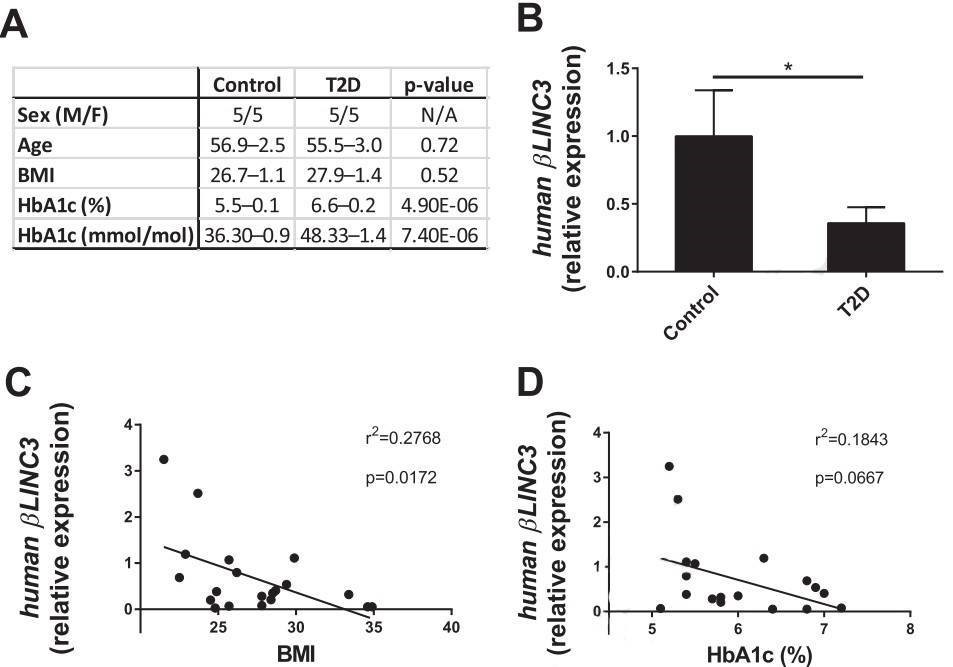 Figure 5. βlinc3 expression is decreased in islets from type 2 diabetes donors.
Figure 5. βlinc3 expression is decreased in islets from type 2 diabetes donors.
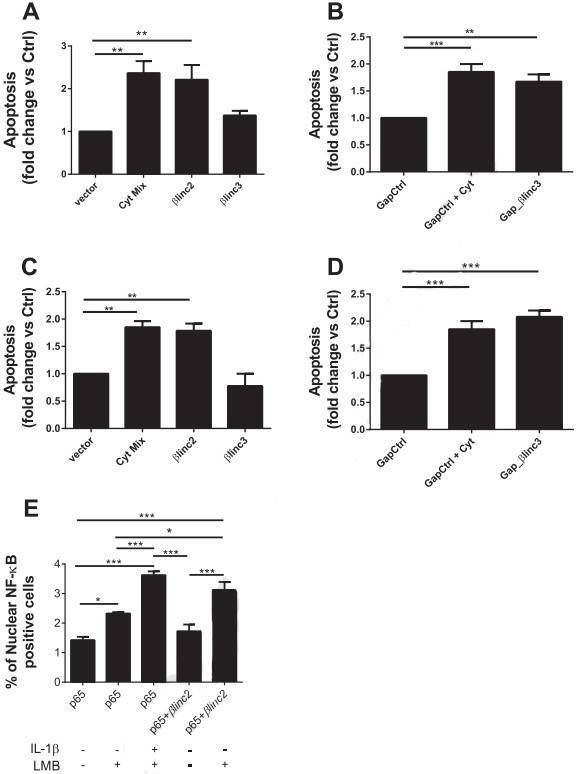 Figure 6. Overexpression and downregulation of the βlinc2 and βlinc3 promotes apoptosis in MIN6 β-cells. (A) MIN6 cells were transfected with a control vector or plasmids to induce the overexpression of βlinc2 or βlinc3. (B) The cells were transfected with a control gapmer or a gapmer to knockdown βlinc3. (C & D) are the repeated experiment in dispersed mouse islet cells. (E) MIN6 cells were transfected with a plasmid expressing GFP-tagged p65.
Figure 6. Overexpression and downregulation of the βlinc2 and βlinc3 promotes apoptosis in MIN6 β-cells. (A) MIN6 cells were transfected with a control vector or plasmids to induce the overexpression of βlinc2 or βlinc3. (B) The cells were transfected with a control gapmer or a gapmer to knockdown βlinc3. (C & D) are the repeated experiment in dispersed mouse islet cells. (E) MIN6 cells were transfected with a plasmid expressing GFP-tagged p65.
Conclusion
This paper identified a large number of novel lncRNAs. At least two of them can affect the survival of β-cells and may contribute to glucolipotoxic-mediated β-cells and the manifestation and progression of T2D. LncRNA therefore may provide the ideal targets for diabetes prevention and treatment.
Reference:
- Motterle A, Gattesco S, Peyot M L, et al. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Molecular metabolism, 2017, 6(11): 1407-1418.
Here are some publications that have been successfully published using our services or other related services:
Restriction endonuclease cleavage of phage DNA enables resuscitation from Cas13-induced bacterial dormancy
Journal: Nature microbiology
Year: 2023
IL-4 drives exhaustion of CD8+ CART cells
Journal: Nature Communications
Year: 2024
High-Fat Diets Fed during Pregnancy Cause Changes to Pancreatic Tissue DNA Methylation and Protein Expression in the Offspring: A Multi-Omics Approach
Journal: International Journal of Molecular Sciences
Year: 2024
KMT2A associates with PHF5A-PHF14-HMG20A-RAI1 subcomplex in pancreatic cancer stem cells and epigenetically regulates their characteristics
Journal: Nature communications
Year: 2023
Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch
Journal: Science Advances
Year: 2024
Genomic imprinting-like monoallelic paternal expression determines sex of channel catfish
Journal: Science Advances
Year: 2022
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
