The CDCap™ Comprehensive Cancer Panel is a hybridization capture-based targeted sequencing assay designed for comprehensive cancer genomic profiling. It detects 641 cancer-related genes, covering loci associated with targeted therapeutic mechanisms, chemotherapeutic agent toxicity/sensitivity, immune pathways, and hereditary cancer predisposition. This panel supports precision oncology research, combination therapeutic strategy investigation, and research cohort selection by integrating biomarkers such as TMB, MSI, MMR, alongside PD-1 pathway regulators and hyperprogression-associated genes for immunotherapy response mechanisms analysis.
Features
Key Features & Advantages
Comprehensive Guidance for Precision Therapy
· Evaluates genes linked to targeted/chemotherapy efficacy, immune pathways, and hereditary predisposition.
Multidimensional Prediction of PD-1 Inhibitor Response
· evaluates MSI, TMB, and MMR to comprehensively predict immunotherapy efficacy.
Optimized for Liquid Biopsy
· Streamlined panel design with UMI tagging enhances sensitivity and accuracy for low-input cfDNA detection.
Data
Data
1. Exceptional Correlation: WES and TMB Panel Results Align with High Precision(R² ≥ 0.9966)
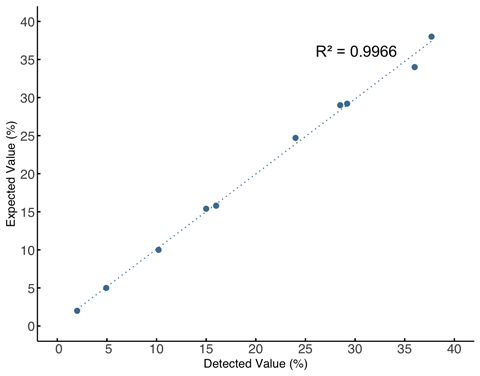
2. High Sensitivity and Accuracy in Mutation Detection
Table 1. Mutation Detection Results for Positive Standards
| Gene | Variant | Expected Allelic Frequency | Reported allelic frequency - 50 ng | Reported allelic frequency - 100 ng |
| ---- | ---- | ---- | ---- | ---- |
| EGFR | L858R | 1% | 0.95% | 1.50% |
| KRAS | A146T | 1% | 1.02% | 0.87% |
| NRAS | Q61K | 1% | 0.84% | 1.23% |
| EGFR | T790M | 2% | 1.61% | 2.11% |
| EGFR | ΔE746_A750 | 1.58% | 1.40% | 1.36% |
| FLT3 | ΔI836 | 2% | 2.81% | 1.94% |
| KIT | D816V | 2% | 2.80% | 2.07% |
| KRAS | G12D | 2% | 1.32% | 2.33% |
| EGFR | V769_D770 insASV | 3% | 1.80% | 1.02% |
| EGFR | G719S | 4% | 3.46% | 3.33% |
| KRAS | G13D | 4% | 2.94% | 4.87% |
| EML4-ALK | Fusion V3 | 5% | 3.50% | 3.61% |
| CD74-ROS1 | Fusion | 6% | 2.37% | 1.61% |
| BRAF | V600E | 7% | 5.02% | 5.75% |
| PIK3CA | H1047R | 7% | 7.28% | 5.83% |
| ERBB2 | Amplification | 6 copies | 6 copies | 6 copies |
Table 2. SNV Detection Results for 5% FFPE Standards
| Gene | Variant | Expected Allelic Frequency | Reported allelic frequency - 50 ng | Reported allelic frequency - 100 ng | Reported allelic frequency - 200 ng |
| ---- | ---- | ---- | ---- | ---- | ---- |
| EGFR | L858R | 5% | 2.48% | 6.51% | 4.29% |
| EGFR | T790M | 5% | 2.64% | 3.64% | 4.30% |
| EGFR | ΔE746_A750 | 5% | 2.11% | 3.17% | 3.37% |
| KRAS | A146T | 5% | 2.60% | 3.78% | 3.88% |
| KRAS | G12D | 5% | 1.78% | 3.19% | 3.77% |
| KRAS | G13D | 5% | 1.51% | 5.57% | 5.93% |
| NRAS | Q61K | 5% | 3.36% | 5.06% | 5.11% |
| PIK3CA | E545K | 5% | 1.39% | 4.12% | 5.73% |
All mutations were successfully detected in tumor gDNA and FFPE standards, with reported allelic frequencies closely matching theoretical values.
3. UMI Tagging Enhances Mutation Detection Rate
Table 3. Mutation Detection Results for cfDNA Reference Standards
| Gene | Variant | Expected Allelic Frequency | Reported allelic frequency |
| ---- | ---- | ---- | ---- |
| EGFR | L858R | 0.50% | 0.19% |
| EGFR | T790M | 0.50% | 0.35% |
| EGFR | ΔE746_A750 | 0.50% | 0.27% |
| KRAS | A146T | 0.50% | 0.23% |
| KRAS | G12D | 0.50% | 0.64% |
| KRAS | G13D | 0.50% | 0.76% |
| NRAS | Q61K | 0.50% | 0.44% |
| PIK3CA | E545K | 0.50% | 0.44% |
The use of UMI tagging in cfDNA liquid biopsy significantly reduces background noise, minimizes false-positive mutations, and improves detection accuracy. Libraries were prepared using 15 ng of Horizon 0.5% cfDNA reference standard (HD780) and enriched via liquid-phase hybrid capture with pan-cancer probes.
Gene List
Gene List
Table1: Gene List
| ABCB1 | ABCC3 | ABL1 | ABL2 | ACVR1 | ACVR1B | AGO2 | AKT1 | AKT2 | AKT3 | ALK | ALOX12B |
| AMER1 | ANKRD11 | APC | APEX1 | AR | ARAF | ARFRP1 | ARID1A | ARID1B | ARID2 | ARID5B | ASNS |
| ASXL1 | ASXL2 | ATIC | ATM | ATR | ATRX | AURKA | AURKB | AXIN1 | AXIN2 | AXL | B2M |
| BABAM1 | BAP1 | BARD1 | BBC3 | BCL10 | BCL2 | BCL2L1 | BCL2L11 | BCL2L2 | BCL6 | BCOR | BCORL1 |
| BCR | BIRC3 | BIRC7 | BLM | BMPR1A | BRAF | BRCA1 | BRCA2 | BRD4 | BRIP1 | BTG1 | BTG2 |
| BTK | C11orf30 | C8orf34 | CALR | CARD11 | CARM1 | CASP7 | CASP8 | CBFB | CBL | CBR3 | CCND1 |
| CCND2 | CCND3 | CCNE1 | CD22 | CD274 | CD276 | CD3EAP | CD44 | CD70 | CD79A | CD79B | CDA |
| CDC42 | CDC73 | CDH1 | CDK12 | CDK4 | CDK6 | CDK8 | CDKN1A | CDKN1B | CDKN2A | CDKN2B | CDKN2C |
| CEBPA | CENPA | CHD2 | CHD4 | CHEK1 | CHEK2 | CIC | CREBBP | CRKL | CRLF2 | CSDE1 | CSF1R |
| CSF3R | CTCF | CTLA4 | CTNNA1 | CTNNB1 | CTTN | CUL3 | CUL4A | CXCR4 | CYLD | CYP17A1 | CYP19A1 |
| CYP1B1 | CYP2C8 | CYP2D6 | CYP4B1 | CYSLTR2 | DAXX | DCUN1D1 | DDR1 | DDR2 | DDX43 | DICER1 | DIS3 |
| DNAJB1 | DNMT1 | DNMT3A | DNMT3B | DOT1L | DPYD | DROSHA | DUSP4 | DYNC2H1 | E2F3 | EED | EGFL7 |
| EGFR | EIF1AX | EIF4A2 | EIF4E | ELF3 | EP300 | EPAS1 | EPCAM | EPHA2 | EPHA3 | EPHA5 | EPHA7 |
| EPHB1 | EPHB4 | ERBB2 | ERBB3 | ERBB4 | ERCC1 | ERCC2 | ERCC3 | ERCC4 | ERCC5 | ERF | ERG |
| ERRFI1 | ESR1 | ESR2 | ETV1 | ETV6 | EWSR1 | EXT1 | EZH1 | EZH2 | FADD | FAM175A | FAM46C |
| FAM58A | FANCA | FANCC | FANCD2 | FANCE | FANCF | FANCG | FANCL | FANCM | FAS | FAT1 | FAT3 |
| FBXW7 | FCGR2A | FCGR3A | FGF10 | FGF12 | FGF14 | FGF19 | FGF23 | FGF3 | FGF4 | FGF6 | FGFR1 |
| FGFR2 | FGFR3 | FGFR4 | FH | FLCN | FLT1 | FLT3 | FLT4 | FOXA1 | FOXL2 | FOXO1 | FOXP1 |
| FRS2 | FSHR | FUBP1 | FYN | GAB2 | GABRA6 | GALNT12 | GATA1 | GATA2 | GATA3 | GATA4 | GATA6 |
| GGH | GID4 | GLI1 | GNA11 | GNA13 | GNAQ | GNAS | GPR124 | GPS2 | GREM1 | GRIN2A | GRM3 |
| GSK3B | GSTA1 | GSTM1 | GSTP1 | H3F3A | H3F3B | H3F3C | HAS3 | HDAC1 | HDAC6 | HGF | HIST1H1C |
| HIST1H2BD | HIST1H3A | HIST1H3B | HIST1H3C | HIST1H3D | HIST1H3E | HIST1H3F | HIST1H3G | HIST1H3H | HIST1H3I | HIST1H3J | HIST2H3C |
| HIST2H3D | HIST3H3 | HLA - A | HLA - B | HMMR | HNF1A | HOXB13 | HRAS | HSD3B1 | HSP90AA1 | HSPB1 | ICOSLG |
| ID3 | IDH1 | IDH2 | IFNGR1 | IGF1 | IGF1R | IGF2 | IKBKE | IKZF1 | IL10 | IL1A | IL4 |
| IL7R | IL8 | INHA | INHBA | INPP4A | INPP4B | INPPL1 | INSR | IRF2 | IRF4 | IRS1 | IRS2 |
| JAK1 | JAK2 | JAK3 | JUN | KAT6A | KDM3B | KDM5A | KDM5C | KDM6A | KDR | KEAP1 | KEL |
| KIT | KLF4 | KLHL6 | KMT2A | KMT2B | KMT2C | KMT2D | KNSTRN | KRAS | LATS1 | LATS2 | LIG4 |
| LIMK1 | LIN28B | LMO1 | LRP1B | LTK | LYN | LZTR1 | MAF | MAGI2 | MALT1 | MAP2K1 | MAP2K2 |
| MAP2K4 | MAP3K1 | MAP3K13 | MAP3K14 | MAPK1 | MAPK3 | MAPKAP1 | MAX | MCL1 | MDC1 | MDM2 | MDM4 |
| MECOM | MED12 | MEF2B | MEN1 | MERTK | MET | MGA | MGMT | MITF | MKNK1 | MLH1 | MLH3 |
| MPL | MRE11A | MSH2 | MSH3 | MSH6 | MSI1 | MSI2 | MST1 | MST1R | MTAP | MTHFR | MTOR |
| MTRR | MUTYH | MXI1 | MYC | MYCL | MYCN | MYD88 | MYO3B | MYOD1 | NBN | NCOA3 | NCOR1 |
| NDRG1 | NEGR1 | NEIL1 | NF1 | NF2 | NFE2L2 | NFKBIA | NKX2 - 1 | NKX3 - 1 | NOS2 | NOTCH1 | NOTCH2 |
| NOTCH3 | NOTCH4 | NPM1 | NQO1 | NQO2 | NRAS | NSD1 | NT5C2 | NTHL1 | NTRK1 | NTRK2 | NTRK3 |
| NUF2 | NUP93 | OPRM1 | P2RY8 | PAK1 | PAK3 | PAK7 | PALB2 | PARK2 | PARP1 | PARP2 | PARP3 |
| PAX5 | PBRM1 | PDCD1 | PDCD1LG2 | PDGFRA | PDGFRB | PDK1 | PDPK1 | PGR | PHB | PHOX2B | PIK3C2B |
| PIK3C2G | PIK3C3 | PIK3CA | PIK3CB | PIK3CD | PIK3CG | PIK3R1 | PIK3R2 | PIK3R3 | PIM1 | PLAT | PLCG2 |
| PLK2 | PMAIP1 | PMS1 | PMS2 | PNRC1 | POLDI | POLE | PON1 | PPARG | PPM1D | PPP2R1A | PPP2R2A |
| PPP4R2 | PPP6C | PRDM1 | PRDM14 | PREX2 | PRKAA1 | PRKAR1A | PRKCI | PRKD1 | PRKDC | PRSS8 | PTCH1 |
| PTEN | PTP4A1 | PTPN11 | PTPRD | PTPRO | PTPRS | PTPRT | QKI | RAB35 | RAC1 | RAC2 | RAD21 |
| RAD50 | RAD51 | RAD51B | RAD51C | RAD51D | RAD52 | RAD54L | RAF1 | RANBP2 | RARA | RASA1 | RB1 |
| RBM10 | RECQL | RECQL4 | REL | RET | RFWD2 | RHBDF2 | RHEB | RHOA | RICTOR | RIT1 | RNF43 |
| ROS1 | RPS6KA4 | RPS6KB2 | RPTOR | RRAGC | RRAS | RRAS2 | RRM1 | RSF1 | RTEL1 | RUNX1 | RUNX1T1 |
| RXRA | RYBP | SDHA | SDHAF2 | SDHB | SDHC | SDDH | SEMA3C | SENSN1 | SENSN2 | SESN3 | SETD2 |
| SETD8 | SYF3B1 | SGK1 | SH2B3 | SH2D1A | SHOC2 | SHQ1 | SLCO1B1 | SLCO1B3 | SLIT1 | SLIT2 | SLX4 |
| SMAD2 | SMAD3 | SMAD4 | SMARCA4 | SMARCB1 | SMARCD1 | SMO | SMYD3 | SNCAIP | SOCS1 | SOD2 | SOS1 |
| SOX10 | SOX17 | SOX2 | SOX4 | SOX9 | SPEN | SPOP | SPRED1 | SPTAl | SRC | SRSF2 | STAG2 |
| STAT3 | STAT4 | STAT5A | STAT5B | STK11 | STK19 | STK40 | SUFU | SUZ12 | SYK | TAF1 | TAP1 |
| TAP2 | TBX3 | TCEB1 | TCF3 | TCF7L2 | TDG | TEK | TERC | TERT | TET1 | TET2 | TGFB1 |
| TGFBR1 | TGFBR2 | TIPARP | TLR2 | TMEM127 | TMPRSS2 | TNF | TNFAIP3 | TNFRSF14 | TNFSF11 | TOP1 | TOP2A |
| TP53 | TP53BP1 | TP63 | TRAF2 | TRAF7 | TRRAP | TSC1 | TSC2 | TSHR | TSHZ2 | TSHZ3 | TTF1 |
| TXN | TXNRD2 | TYMS | TYRO3 | U2AF1 | UGT1A1 | UGT1A4 | UMPS | UPF1 | VEGFA | VHL | VTCN1 |
| WHSC1 | WHSC1L1 | WISP3 | WT1 | WWTR1 | XIAP | XPC | XPO1 | XRCC1 | XRCC2 | XRCC3 | YAP1 |
| YES1 | ZBTB2 | ZFHX3 | ZNF217 | ZNF703 | | | | | | | |
| NOTCH2 | NTRK1 | ALK | MSH2 | RAF1 | ETV5 | FGFR3 | SLC34A2 | PDGFRA | KIT | CD74 | ROS1 |
| MYB | EZR | EGFR | BRAF | FGFR1 | MYC | NTRK2 | RET | FGFR2 | KMT2A | ETV6 | BRCA2 |
| NUTM1 | RARA | BRCA1 | ETV4 | SDC4 | TMPRSS2 | BCR | EWSR1 | BCL2 | RSPO2 | MET | ETV1 |
| PPARγ | TERT | | | | | | | | | | |
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.