Single nucleotide polymorphism (SNP) occurs relatively frequently in the human genome, with an average of one polymorphic site per 1,000 bases, and is the third generation of genetic markers following microsatellites. Some SNPs also affect gene function, which can lead to changes in biological traits and even cause disease.
CD Genomics provides SNaPshot Multiplex System for SNP detection. The system is based on the principle of fluorescently labeled single base extension. Mainly for medium-throughput SNP testing projects. It is widely used in population genetics research (such as the origin, evolution and migration of organisms) and disease-related genes. In addition, it also plays an important role in pharmacogenomics, diagnostics, and biomedical research.
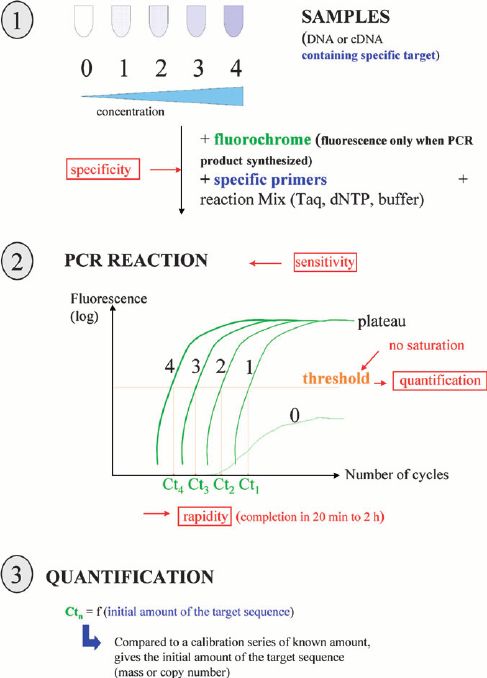
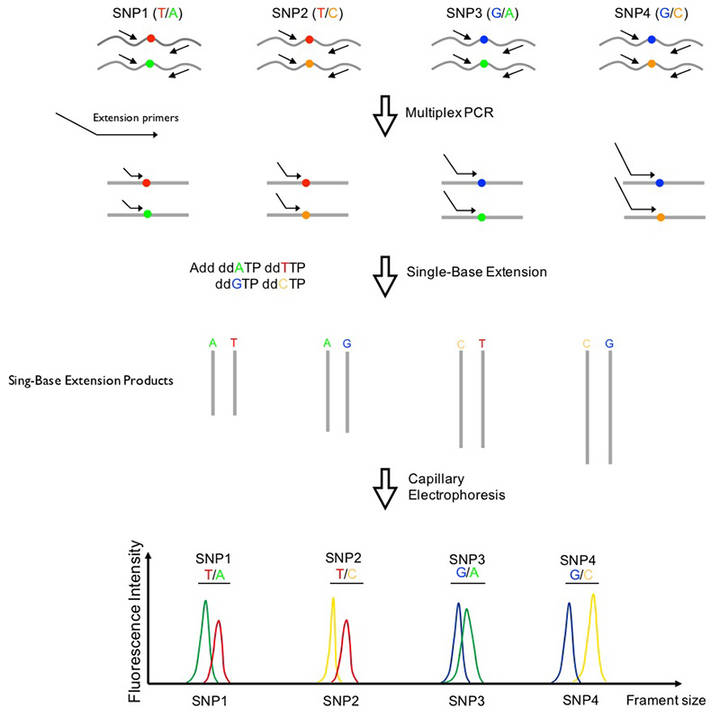
The principle and process of SNaPshot Multiplex System
First, the primers are designed according to the known locations. The primers for different sites are designed into different lengths, and then the SNaPshot reaction is performed. In a reaction system containing a sequenced enzyme, four fluorescently labeled ddNTPs, primers of different lengths next to the 5 ′ end of the polymorphic site, and a PCR template, the polymerase extended the primer by one nucleotide and then terminated, adding a ddNTP to its 3 'end. After detection by the ABI sequencer, the type of bases incorporated can be known according to the color of the peaks, thereby determining the genotype of the sample. The SNP site corresponding to the extension product was determined according to the position of the peak shift. This method is mainly used for medium-throughput SNP typing projects, and is usually used for the analysis of 10-30 SNP loci.
Technology platform
ABI PRISM 3700 DNA Analyzer
Advantages of Multiplex SNaPshot service
- High precision, which can be matched with sanger sequencing. This technique can directly obtain the base composition of the site, and the detection results are intuitive.
- It is not limited by the polymorphism of SNP loci, regardless of whether the locus is heterozygous, or inserted or deleted polymorphic, can be detected in a system.
- A contaminated sample can be detected. If the typed peak spectrum of a sample deviates from the normal distribution, it can indicate that the sample may be contaminated or concentrated.
- Medium-throughput: SNaPshot simultaneously interrogates up to 30 SNPs in a single reaction.
- Low cost per SNP.
Workflow of Multiplex SNaPshot service

Sample requirements
- Tissue samples: stored in low-temperature refrigerator at -20℃ or -80℃, transported with liquid nitrogen or dry ice; local or short-distance transport can be ice pack transport. If the material provided is biological material such as fresh tissue or blood cells, please provide enough material to extract more than 2ug of genetic DNA.
- Cultured cells in vitro (≥ 106 cells).
- DNA samples: DNA concentration ≥ 20ng / ul, total DNA ≥ 1ug, OD260/280 between 1.7 and 2.0, without PCR inhibitor. Samples must be shipped in a cold ice pack. FFPE samples can also be received.
- Blood samples: it is required to be stored in anticoagulant tubes or cryopreservation tubes with a volume greater than 2ml. Please transport them in dry ice to ensure that DNA is not degraded.
Deliverables
Peak file (abl. Format), sequence file (seq. Format); formal data analysis report.
Want to know about other Non-NGS gene mutation detection methods?
References:
- Mehta B, Daniel R, Phillips C, et al. Forensically relevant SNaPshot® assays for human DNA SNP analysis: a review. International journal of legal medicine, 2017, 131(1): 21-37.
- Kwong A, Ho J C W, Shin V Y, et al. Rapid detection of BRCA1/2 recurrent mutations in Chinese breast and ovarian cancer patients with multiplex SNaPshot genotyping panels. Oncotarget, 2018, 9(8): 7832.
- Magnin S, Viel E, Baraquin A, et al. A multiplex SNaPshot assay as a rapid method for detecting KRAS and BRAF mutations in advanced colorectal cancers. The Journal of Molecular Diagnostics, 2011, 13(5): 485-492.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources