MassARRAY, A High-Precision and Cost-Effective Technology for Cancer Research
Tumors develop or metastasize from mutations that escaped the elimination of the immune system of the body. Hijacking the cell machinery, tumors can achieve unrestricted growth, progressing to cancer, and multiply in different parts of the body. Huge hurdles in diagnosis and rising numbers of incidences compel pharmaceutical companies to design a growing number of compounds for treatment. A rising trend in oncology nowadays is in the area of personalized medicine which features analysis of the patient's tumor thereby establishing known mutations that are present in the sample. The long excruciating road to diagnosis was conventionally performed through fluorescence in-situ hybridization (FISH) analysis and reverse transcriptase PCR (RT-PCR) to identify mutations although both are prone to artifacts.
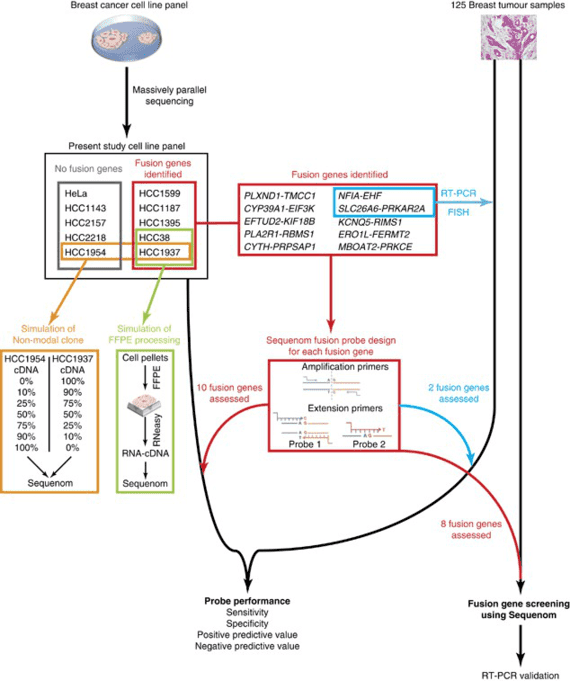 Figure 1. High-throughput detection of fusion genes in cancer using MassARRAY technology. (Lambros, 2011)
Figure 1. High-throughput detection of fusion genes in cancer using MassARRAY technology. (Lambros, 2011)
Relatively recent advancement in the analysis of biological macromolecules is the MassArray based on mass spectrometry using MALDI-TOF (Matrix-Assisted Laser Desorption/Ionization-Time of Flight). It can be used to identify pathogens in clinical microbiology. In the field of oncology, it facilitates accurate identification of a broad spectrum of SNPs, somatic mutations, and also DNA methylation from different sample types. Basically, the MALDI-TOF begins with the emission of laser energy absorbed by the matrix which results in partial vaporization and infliction of minimal fragmentation to the illuminated substrate. Then, ionized samples are electrostatically transferred into the mass spectrometer; enabling detection by the mass-to-charge ratio. Like gel electrophoresis, MassARRAY distinguishes DNA molecules through ionization resulting in a variable known as the time-of-flight which directly translates to its molecular mass without the need of additional labeling reagents such as fluorescent techniques.
Particularly, MassARRAY SNP Genotyping involves the amplification of the sequence across the SNP site through PCR using a single extension primer which ensures that the primer extends only one base. The amplified product is then analyzed by time of flight and the SNP is classified based on the molecular weight difference of base; the allele frequency of the SNP site could also be calculated. This platform is actively being used for the diagnosis of colorectal and lung cancer in patients.
With reference to its counterparts, MassARRAY offers higher precision, reproducibility, sensitivity, and cost-effectiveness. When compared, next-generation sequencing (NGS) may produce more errors than the MassARRAY platform; however, the former could be improved through repetitive sequencing and consolidation of data which may be relatively time-consuming and costly. Moreover, analysis and interpretation of data from the NGS platform is a tough hurdle to overcome while MassARRAY provides more straight-forward reports.
References:
- Hrabak J, Bitar I, Papagiannitsis CC. Combination of mass spectrometry and DNA sequencing for detection of antibiotic resistance in diagnostic laboratories. Folia microbiologica. 2020, 65(2): 233-43.
- Basu P, Chandna P, Bamezai RN, et al. MassARRAY spectrometry is more sensitive than PreTect HPV-Proofer and consensus PCR for type-specific detection of high-risk oncogenic human papillomavirus genotypes in cervical cancer. Journal of clinical microbiology. 2011, 49(10).
- Lambros MB, Wilkerson PM, Natrajan R, et al. High-throughput detection of fusion genes in cancer using the Sequenom MassARRAY platform. Laboratory investigation. 2011, 91(10).
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
 Figure 1. High-throughput detection of fusion genes in cancer using MassARRAY technology. (Lambros, 2011)
Figure 1. High-throughput detection of fusion genes in cancer using MassARRAY technology. (Lambros, 2011)