Introduction
Melanoma develops from the pigment-containing cells in the skin which is known as melanocytes. Melanoma usually occurs on normal skin, but can also occur in the mouth, intestines or in the eyes. It can be characterized as sudden appearance or rapid growth of the numerous pigmented macules, deepening in color, surrounded by stellate nodules or pigmented rings, local pain, infection, ulcers or bleeding and enlarged lymph nodes. Prolonged exposure of skin cells to the ultraviolet light can cause damage to genetic material in skin cells, which may result in uncontrolled cell growth and development of cancer. The pigment can protect skin cell and block UV radiation from damaging DNA. Due to UV radiation, cutaneous melanomas show a markedly elevated base mutation rate compared to other solid tumors because of the increasing cytidine to thymidine (C > T) transitions.
Disease-related gene description
The mutations in NRAS result in abnormally sustained activation of downstream genes such as Raf kinase. And the mutations in BRAF gene can affect tumor proliferation, growth and differentiation. Pathogenic variant in NRAS, BRAF and other common genetic aberrations, such as PTEN and TP53 genes may increase the risk of malignant melanoma. The inactivation of NF1, which is a negative regulator of RAS signaling, may activate aberrant mitogen-activated protein kinase (MAPK) pathway. CDKN2A is a suppressor gene for melanoma tumor encoding two tumor suppressor proteins by variable splicing. One of them is p16INK4a, a cyclin-dependent kinase inhibitor, and the other is p14ARF, which activates p53 through inhibiting the expression of MDM2. In addition to these well-known cancer genes, new candidates (PPP6C, RAC1, SNX31, TACC1, STK19 and ARID2) were also reported to be the significantly mutated melanoma genes.
Our custom melanoma panel platform offers a comprehensive melanoma gene panel, from which you can choose the genes only associated with your researches. Amplicon sequencing technology that enables deep sequencing at high coverage levels is provided. Our customized melanoma panel can be applied to efficiently discover, validate and screen genetic variants among the melanoma genes to help research the new therapeutic strategies of melanoma biology.
Custom melanoma panel offers but are not limited to:
-
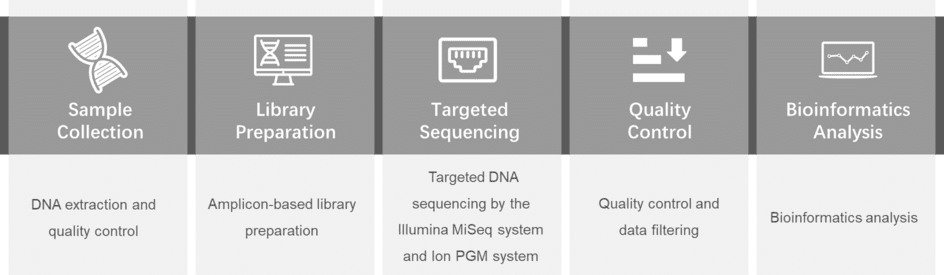
We provide automation and high-throughput targeted amplicon sequencing technology with the Illumina MiSeq system/Ion PGM system.
-
Strict quality control throughout the pipeline workflow ensures the accuracy and repeatability of the sequencing.
-
Every detected genetic variant will be further validated to ensure the validity of results.
-
Custom panel content is designed to keep up with the frontiers from current literature about melanoma panel to target all relevant regions.
-
You can choose the panel content from our melanoma cancer panel library or discuss your customized melanoma requirements, then we can provide you with your own panel.
-
Precision bioinformatics pipelines ensure superior analytical performance.
Choose the genes that suit you from the melanoma gene list
| ARID2 |
BAP1 |
BRAF |
BRCA2 |
| CDK4 |
CDKN2A |
CHEK2 |
ERBB4 |
| GNAQ |
GNA11 |
KIT |
MC1R |
| MITF |
MUTYH |
NRAS |
NF1 |
| POT1 |
PPP6C |
PREX2 |
PTEN |
| RAC1 |
RB1 |
SLC45A2 |
SNX31 |
| STK19 |
TACC1 |
TP53 |
TYR |
Specimen requirements of our custom melanoma panel
- Specimen: blood, saliva or extracted DNA (we do not accept DNA samples isolated from FFPE tissue).
- Volume: 2-5 mL blood, 2 mL saliva 3ug DNA.
- Collection: blood is collected by routine blood collection and saliva is collected by saliva collection kits (kits are available upon request). DNA samples are stored in TE buffer or equivalent.
- Container: lavender-top (EDTA) tube or yellow-top (ACD) tube.
- Storage/transport temperature: room temperature.
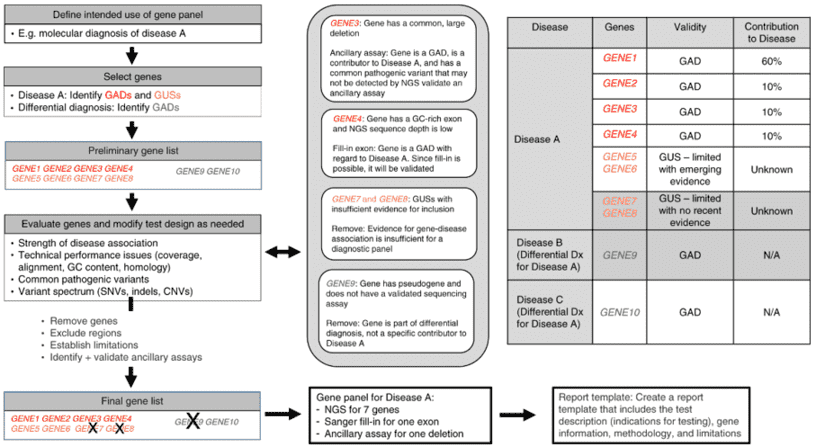
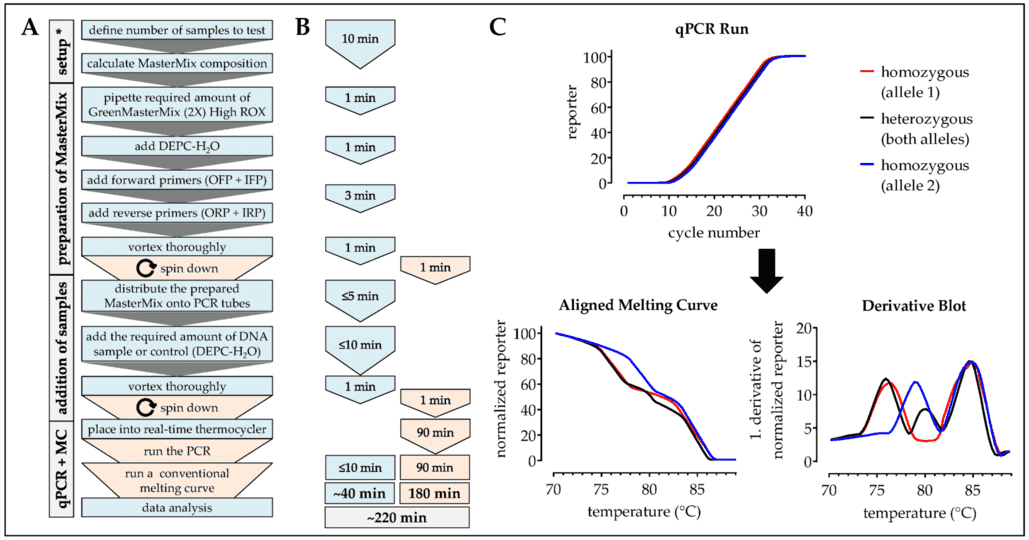
Gene panel workflow
For more information about the Custom Melanoma Panel or need other amplification requirements, please contact us.
References:
- De Unamuno Bustos B, et al. Towards personalized medicine in melanoma: implementation of a clinical next-generation sequencing panel. Scientific reports, 2017, 7(1): 495.
- Kelleher F C, McArthur G A. Targeting NRAS in melanoma. The Cancer Journal, 2012, 18(2): 132-136.
- Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature, 2010, 467(7315): 596.
- Hodis E, et al. A landscape of driver mutations in melanoma. Cell, 2012, 150(2): 251-263.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources