- Resource
- Applications
- Metagenomic Next-Generation Sequencing (mNGS) in Respiratory and Lung Pathogen and Disease Research
Metagenomic Next-Generation Sequencing (mNGS) in Respiratory and Lung Pathogen and Disease Research
What is metagenomic next-generation sequencing (mNGS)?
Metagenomic next-generation sequencing (mNGS) is a nucleic acid detection-based microbial identification technology that has emerged in recent years and is widely used because of its non-predictive and high-throughput advantages. Since the identification of infectious microorganisms is complex and the identification of infections and colonization is difficult, customized pathogen-targeted sequencing kits based on mNGS technology can detect unknown or non-predictable pathogens at once. With a wider detection range, it can be used as a basis for pathogen research in combination with high-throughput sequencing technology.
| Advantages |
Application |
- High sensitivity: allows detection of low copy numbers of pathogens
- High specificity: detects multiple pathogens, subtypes and new species at once
- More comprehensive information: traditional methods can only determine the presence or absence of pathogens, mNGS can provide more detailed information such as drug susceptibility of the pathogen, genomic information of the host, etc.
- High throughput: multiple samples can be analyzed simultaneously, improving detection efficiency
|
- Diagnosis method development: detect infections and diseases with unclear pathogens or unclear sources of infection
- Epidemic surveillance: monitor the spread of pathogens in the population and quickly detect changes in pathogen infections so that corresponding prevention and control strategies can be developed
- Food safety: detect pathogen contamination in food
- Environmental surveillance: monitor the distribution of pathogens in the environment
|
mNGS and pulmonary infectious diseases
Pulmonary infections are a major cause of mortality and morbidity worldwide, and are defined as substantive lung infections including bronchial, alveolar and interstitial infections caused by various pathogens including bacteria, fungi and viruses. Metagenomic next generation sequencing (mNGS) technology is an emerging pathogen detection method that can effectively identify pathogens by sequencing all microorganisms in a specimen. It has the advantages of wide coverage, relatively short cycle time and low impact by early application of antibiotics compared with traditional detection methods.
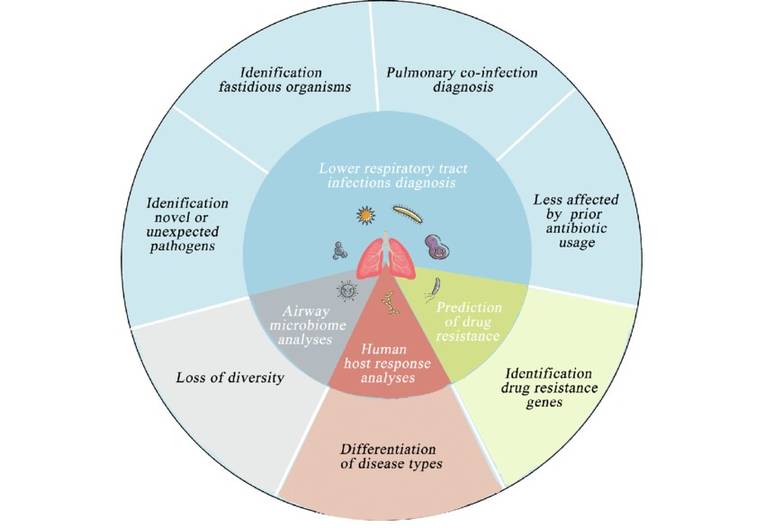 Applications of mNGS in the area of lower respiratory tract infections.
Applications of mNGS in the area of lower respiratory tract infections.
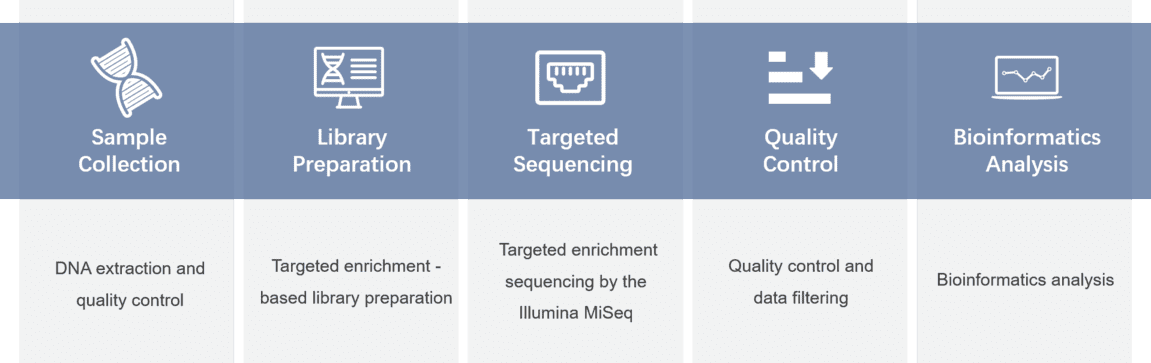
For example, CD Genomics offers a fast and convenient pathogen detection solution for a variety of types and species using advanced target enrichment NGS technology. Our amplicon and hybrid capture-based technologies ensure the detection of low copy number pathogens, while sequencing technology allows the detection of multiple pathogens, as well as subtypes and novel species, in a single experiment.
Application of mNGS in bacteria
Bacteria occupy an important position in the pathogens of pulmonary infections, and the most common detection method in clinical practice is bacterial culture of secretions and other specimens, but the culture results are generally only for common bacteria, and the results are poor for special and rare bacteria. Numerous studies have shown that the positive rate of mNGS for pathogenic bacteria is higher than that of the traditional culture method, making up for the shortcomings of traditional bacterial culture. mNGS is not only superior in the detection of common bacteria, but also has greater value for the detection of rare bacteria.
Our Bacteria NGS Panels based on amplicon sequencing on Illumina platforms, enable to detection of the 16S rRNA variable regions (V1-V9) in a fast, accurate and safe workflow. We offer solutions specifically for lung and respiratory bacterial characterization, and our sequencing and bioinformatic analysis teams can provide additional customization and analysis services.
Application of mNGS in fungi
The prevalence of pulmonary fungal diseases has been increasing significantly in recent years as the number of high-risk groups requiring immunosuppressive drugs for medical conditions and other reasons has increased. Most of the fungi infecting human lungs are opportunistic pathogens, and traditional detection methods are time-consuming.
The ITS regions (ITS1 and ITS2 regions) are usually selected for sequencing because they are under less pressure from natural selection during evolution, and the ITS regions evolve rapidly and exhibit sequence polymorphism in most fungi. Our Fungi mNGS Panels are designed to detect ITS regions as well as antibiotic-resistance gene clusters for subgenus-level identification and functional analysis.
Application of mNGS in viruses
Lung infections caused by viruses are not uncommon in clinical work, such as rhinovirus, adenovirus, coronavirus and respiratory syncytial virus. Traditional virus detection methods include serological tests (antigen and antibody tests) and nucleic acid tests, but these tests target specific viruses and have a small viral coverage, whereas mNGS tests can detect almost all pathogens in a sample and therefore have a significant advantage in pathogen detection.
With advantages such as unbiased genome-wide coverage (over 99%) and short hand-on times, our viral mNGS kits allow for the single detection of dozens of common respiratory viruses and are suitable for viral evolutionary analysis, detection and resistance studies.
Application of mNGS to specific pathogens
Mycoplasma and Chlamydia, as atypical pathogens, are also common causative agents in lung infections, especially mycoplasma often causes lung infections in children and young adults. In addition, mNGS has also been applied to chlamydial infections, and studies have reported cases of Chlamydia psittaci not detected by traditional methods but detected by mNGS technology, suggesting the potential advantage of mNGS in the detection of Chlamydia psittaci pneumonia.
Detection of mixed infections in lung infections
Although mixed infections are quite common in clinical practice, it is very difficult to confirm the diagnosis of pathogens, for example, some pathogens such as viruses, parasites, fungi, etc. Laboratory techniques cannot be adapted to clinical needs, and cultures of body fluids at non-sterile sites suggesting multiple bacterial growth do not indicate the existence of a true mixed infection, and on the contrary, sterile growth or single bacterial growth cannot exclude mixed infections. mNGS can detect multiple pathogens such as bacteria, fungi, viruses and parasites at one time, which can significantly improve the detection rate of each pathogen in multiple mixed infection patterns and can provide direction for subsequent clinical diagnosis and treatment due to its non-predetermined and non-preferential characteristics.
Metagenomic technology can also predict antimicrobial drug resistance, identify virulence genes, and be of outstanding value for strain typing, especially in detecting pathogens that are not easily cultivated. Currently, this technology uses a variety of clinical samples such as respiratory samples, blood, cerebrospinal fluid, human tissues and stool to detect infectious pathogens, and the detection and analysis time can be controlled to less than 24 h, with the fastest being 6 h.
mNGS does not require pre-defined target pathogens, does not require specific primers, is not affected by genetic mutations, has a short detection period, broad detection range, high sensitivity, and mNGS can identify pathogens for typing and virulence gene detection. The detection rate of mNGS in treatment is less affected by antibiotics than conventional detection methods, and is less likely to result in false negative pathogen detection due to early empirical use of antibiotics, and the results of mNGS can guide the use of antibiotics. In addition, mNGS has made some clinical progress for the detection of drug resistance genes. mNGS can be used to evaluate the diversity and resistance of drug resistance genes by sequencing the genomes of microorganisms, which in turn has a guiding significance for the discovery of new drug resistance targets and also has an important potential value for the development of new sensitive antibiotics for drug-resistant flora.
Reference:
- Diao Z, Han D, Zhang R, et al. Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections. Journal of Advanced Research, 2022, 38: 201-212.
* For Research Use Only. Not for use in diagnostic procedures.
 Applications of mNGS in the area of lower respiratory tract infections.
Applications of mNGS in the area of lower respiratory tract infections.