Introduction
Acute myeloid leukemia (AML) is a malignant disease of myeloid hematopoietic stem/progenitor cells. The main feature of AML is abnormal proliferation of primitive and naive myeloid cells in bone marrow and peripheral blood. This disease has high incidence in new-born and the aged, and accounts for 30% of pediatric leukemia. Children with AML are similar to adults (< 50) in molecular biology changes and chemotherapy responses. AML in infants and young children is prone to extramedullary leukemia. The clinical manifestations are anemia, hemorrhage, infection and fever, organ infiltration and metabolic abnormalities. In most cases, the condition is acute and the prognosis is dangerous. If not treated in time, it can often be life-threatening. Therefore, the study of the mechanism of AML development at the gene level is the basis for exploring AML.
Disease-related gene description
Acute myeloid leukemia gene 1, 2 and 3 (AML1, AML2, AML3), also known as RUNX1, RUNX3 and RUNX2, are involved in hematopoiesis. The mutations in RUNX1, RUNX3 and RUNX2 are associated with several types of leukemia. Otherwise, the pathogenic variants in some genes such as class III receptor tyrosine kinase (FLT3), DNA methyltransferase 3 alpha (DNMT3A) and CCAAT enhancer binding protein alpha (CEBPA) can cause AML in different ways. FLT3 regulates hematopoiesis. The mutations of FLT3 cause constitutive activation of this receptor, resulting in acute myeloid leukemia and acute lymphoblastic leukemia. DNMT3A is an epigenetic modification that is important for embryonic development, and CEBPA functions in homodimers and also heterodimers with enhancer-binding proteins beta and gama. Mutation of CEBPA is proved associated with acute myeloid leukemia.
To further investigate the mutations in the pathogenesis of AML, the changes in related genes, and the evolution at the genetic level, our platform provides you with a comprehensive AML library and you can choose the gene you specifically need in the library. We provide targeted gene sequencing technology to detect, validate and filter genetic variants among the AML genes. We also provide service to help researchers process the complexities of AML biology quickly.
Custom AML panel offers but are not limited to:
-
You can customize the AML panel through our AML gene library. Or you can contact us with your specific order, then we can draft your own panel.
-
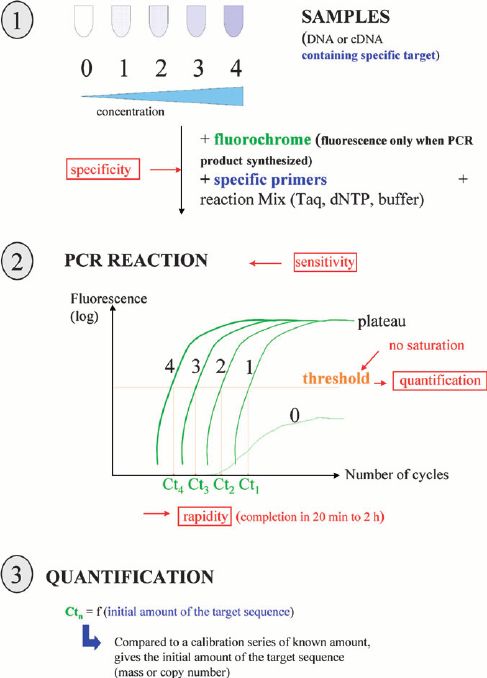
Efficient sequencing technologies with advanced target enrichment methods are offered. Targeted DNA sequencing technology by Ion PGM system/Illumina MiSeq system provides ultra-deep sequencing to target specific genomic regions.
-
Every genetic variant we detect will be further validated to guarantee the accuracy of results.
-
Strict quality control throughout the pipeline workflow ensures the accuracy and repeatability of the sequencing.
-
Our AML gene library will update frequently according to the latest research progress to keep high coverage uniformity.
Choose the genes that suit you from the AML gene list
| ASXL1 |
FAM154B |
IDH2 |
KDR |
NPM1 |
| PRAMEF2 |
C17orf97 |
FAM47A |
JAK1 |
KIT |
| NRAS |
PTPN11 |
CBL |
FAM5C |
JAK2 |
| KRAS |
NTRK3 |
RUNX1 |
CEBPA |
FLRT2 |
| JAK3 |
LRRC4 |
OR13H1 |
TET2 |
DNMT3A |
| FLT3 |
KCNA4 |
MLL3 |
OR8B12 |
TP53 |
| EGFR |
GJB3 |
KCNK13 |
MYC |
P2RY2 |
| TUBA3C |
EZH2 |
IDH1 |
KDM6A |
NF1 |
| PCDHB1 |
WT1 |
|
|
|
Specimen requirements of our custom AML panel
- Specimen: extracted DNA (we do not accept DNA samples isolated from FFPE tissue).
- Volume: 6 μg total DNA.
- Collection: DNA samples are stored in TE buffer or equivalent.
- Container: eppendorf micro test tubes
Gene panel workflow
Need more information about the Custom AML Panel or have other requirements in detail? Please contact us.
References:
- Thakral, G, et al. AML multi-gene panel testing: A review and comparison of two gene panels. Pathology–Research and Practice 2016, S0344033816300152.
- Alphen C, et al. abstract 1820: Phosphoproteomics of a panel of AML cell lines receals oncogenic signaling and candidate drivers. Cancer Research 2015, (15) 1820-1820.
- Cher, C Y, et al. Next-generation sequencing with a myeloid gene panel in core-binding factor AML showed KIT activation loop and TET2 mutations predictive of outcome. Blood Cancer 2016, (7) e442.
- Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood, 2017, 129(4): 424-447.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources