Sequencing Methods for Tumor Profiling and Mutation Load in Cancer Research
A large family of diseases that involver abnormal cell growth with the chance of invading the other body parts are known as cancers. They form a subset of neoplasm, or tumor, which is a group of cells that have undergone unregulated growth. Neoplasms often form a mass, also known as lump, but may also be distributed diffusely. One of the well-known characteristics of cancers is out-of-control cell growth. Increased cell growth means an increased chance of having changes or mutations in the DNA. These mutations may keep on accumulating in the cells and cross the threshold limit, which then may result in abnormal cell division and eventually in cancer.
It is important to understand that cancers are not purely hereditary, or only caused by acquired mutations. Many different types of cancers are caused by a combination of genetic inheritance and the environment. The type of mutation involved in this cause is called a somatic mutation. This is different from germline mutations which are inherited. Mutation load, a type of genetic load that reflects the total genetic burden in a population resulting from accumulated deleterious mutations, can be determined by sequencing. It is a powerful tool in predicting the risk of a person for developing cancer or predicting their response to treatment if they have cancer. The two main focused areas for cancer research are prevention and treatment. In doing so, researchers apply two different approaches, tumor profiling, and mutation load.
Targeted Next-Generation Sequencing
One of the widely used sequencing methods nowadays in getting valuable insight into cancer treatment and prevention is the next-generation sequencing (NGS), specifically using cancer panel that utilizes NGS. This sequencing method has several approaches to cancer research. First is the whole genome sequencing which provides a comprehensive view of the genetic sequence of the samples such as blood, tumor, or other tissues. Second is the targeted NGS which focuses on specific areas of the genome that might specifically more relevant for cancer. Some examples of targeted NGS include exome sequencing and hybrid-capture targeted NGS. Next-generation sequencing can include DNA and RNA sequencing. RNAs are also sequenced as it changes the way protein is expressed which might be a great help in understanding cancer even more.
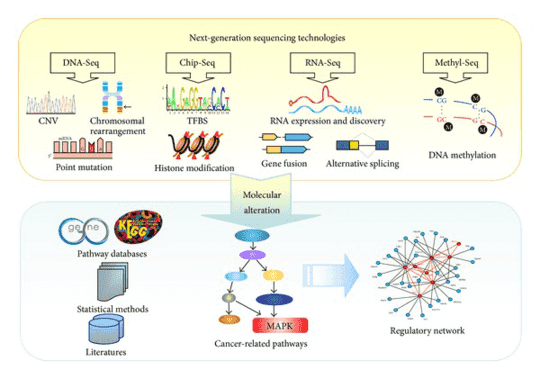 Figure 1. NGS-based pipeline for cancer marker discovery. (Chen, 2013)
Figure 1. NGS-based pipeline for cancer marker discovery. (Chen, 2013)
Whole Genome Sequencing
This sequencing method can provide a base-by-base view of the specific mutations present in given cancer tissue. It enables the discovery of several novel cancer-associated variants, such as copy number changes, structural variants, single nucleotide variants (SNVs), as well as insertiondeletions (indels).
Circulating Tumor DNA (ctDNA) Sequencing Analysis
This sequencing method uses cell-free circulating tumor DNA in detecting cancer since ctDNA can act as a noninvasive cancer biomarker. This offers a potential alternative to invasive tissue biopsies, and noninvasive approach for real-time monitoring of treatment response and identifying candidates for therapy. Recently, translational cancer researchers have been investigating liquid biopsies to detect ctDNA in tumors.
Cancer Exome Sequencing
Exome sequencing is one useful method in evaluating mutation load. This sequencing method provides insight into the protein-coding region of the genes which frequently contain mutations that can affect the progression of the tumor. The content of cancer exome sequencing is possible to expand into the untranslated regions and microRNA binding sites depending on experimental requirements.
Cancer RNA-SEQ
This method involves sequencing of coding regions, also known as whole cancer transcriptome, and can provide crucial information about gene expression changes in tumors. It specifically enables the detection of strand-specific information which is an important component of gene regulation. It can provide a complete orientation that can be used for visualizing the complete view of expression dynamics.
References:
- Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome medicine. 2015, 7(1).
- Chen J, Zhang D, Yan W, et al. Translational bioinformatics for diagnostic and prognostic prediction of prostate cancer in the next-generation sequencing era. BioMed research international. 2013.
- Ku CS, Cooper DN, Roukos DH. Clinical relevance of cancer genome sequencing. World journal of gastroenterology: WJG. 2013, 19(13).
- Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. American journal of clinical pathology. 2011, 136(4).
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
 Figure 1. NGS-based pipeline for cancer marker discovery. (Chen, 2013)
Figure 1. NGS-based pipeline for cancer marker discovery. (Chen, 2013)