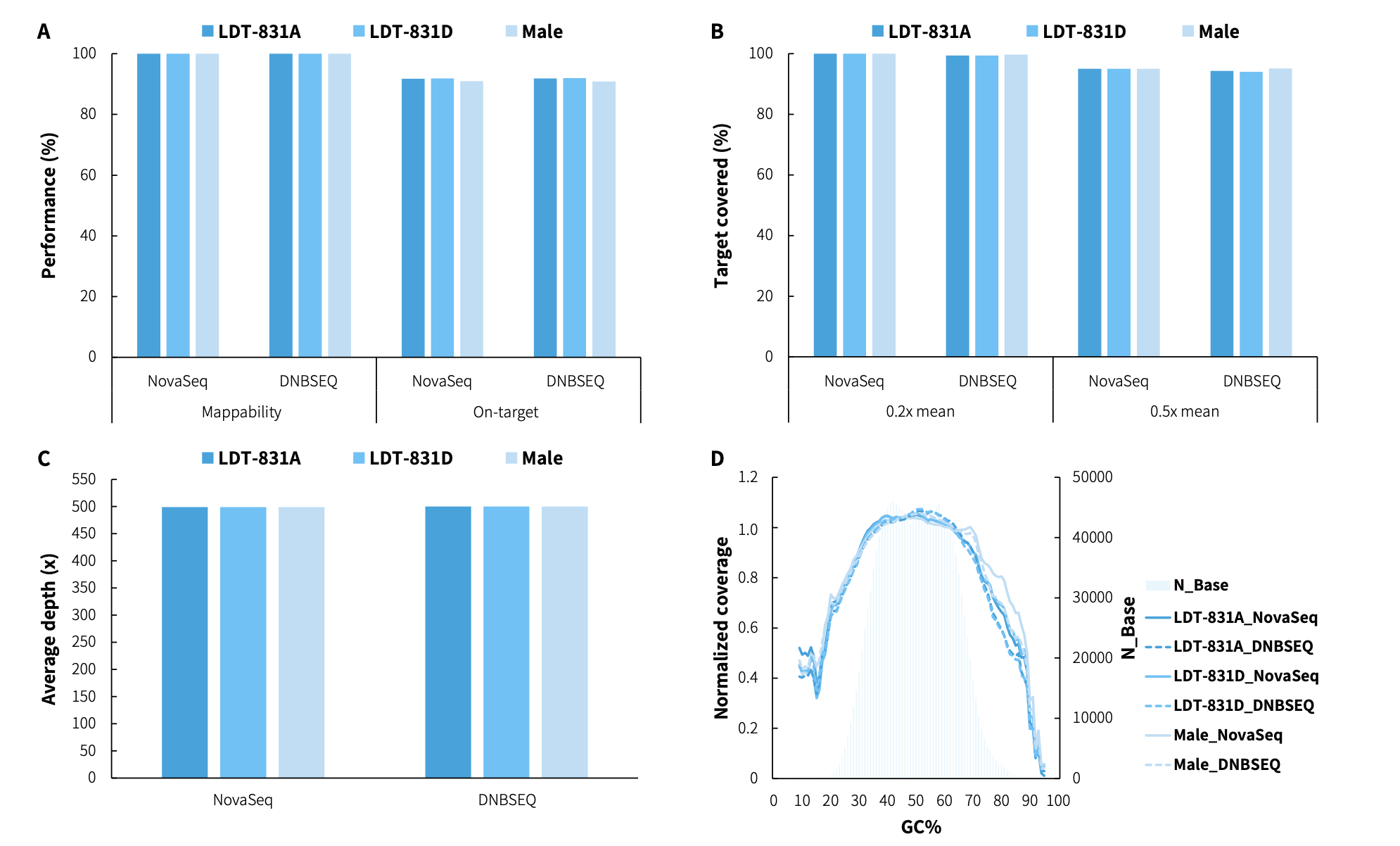
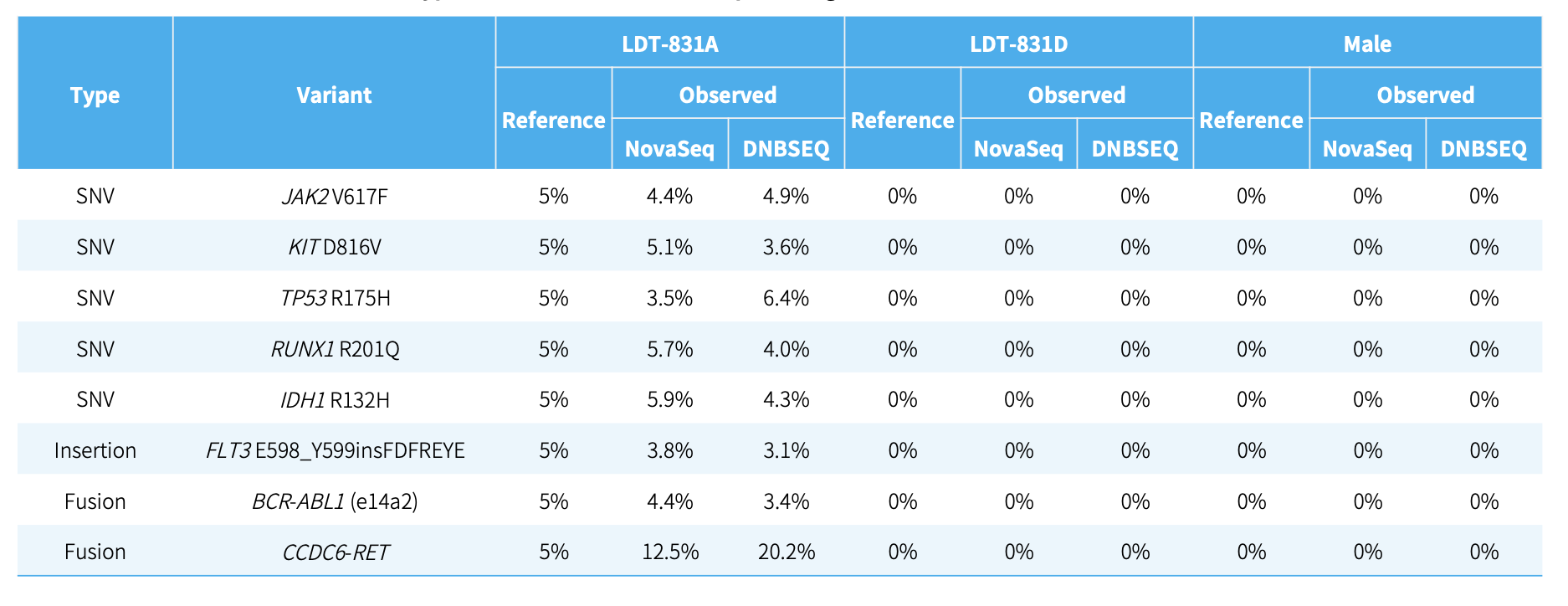
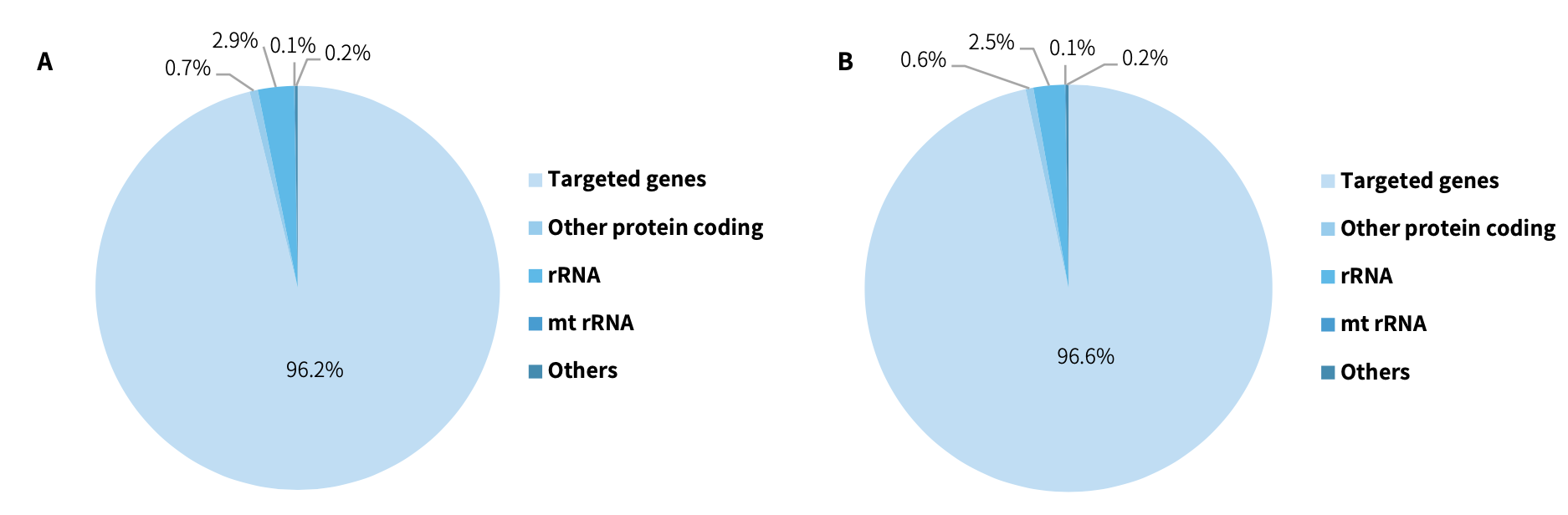
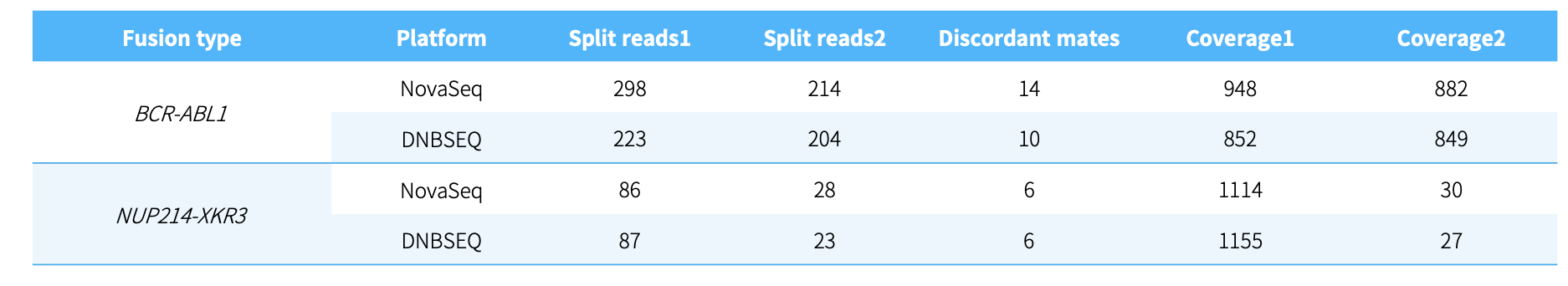
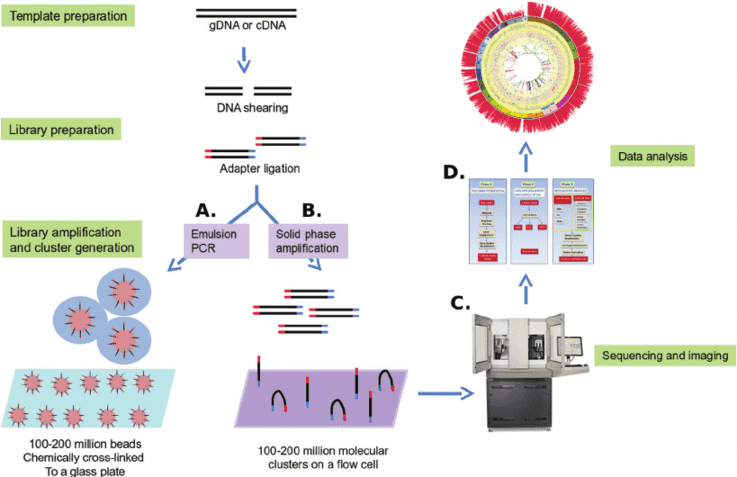
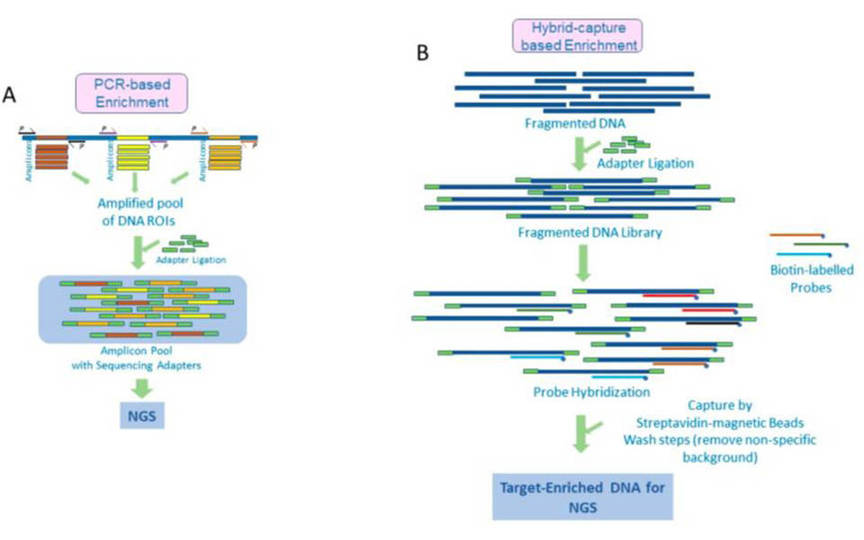
CDCap™ Hema-Tumor Expanded Gene Panel employs a hybridization capture-based dual-omics architecture to comprehensively interrogate 481 clinically relevant genes in hematologic malignancies. This integrated platform enables simultaneous detection of diverse genomic aberrations, including single-nucleotide variants (SNVs), insertions/deletions (Indels), copy number variations (CNVs), and fusion events.
Hema-Tumor DNA Tube
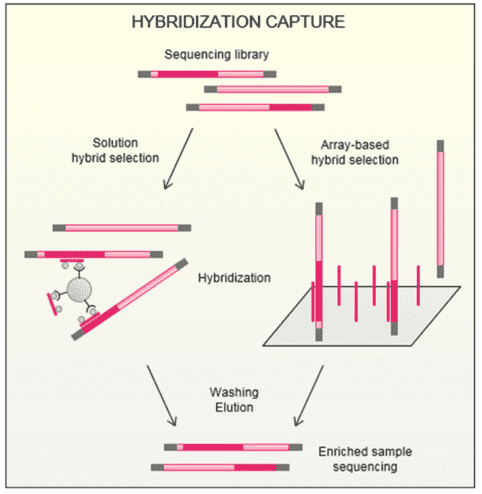
Covers 1.7 Mb of genomic regions, including the full coding regions of 436 genes, partial intronic regions of 12 genes, and the IGH Switch regions for robust mutation analysis.
Hema-Tumor RNA Tube
Targets 146 genes with optimized primer design to accurately detect fusion genes, complementing the DNA workflow for resolving structural variants.
By combining multi-omics approaches with a highly targeted design, the CDCap™ Hema-Tumor Expanded Gene Panel delivers a cost-effective solution for precision analysis, research decision support, and longitudinal tracking of hematologic malignancies.
Features
Key Features & Advantages
Comprehensive Mutation Coverage
· Enables detection of base substitutions, indels, amplifications, and gene fusions across 481 hematologic malignancy-associated genes.
Synergistic Dual Workflow
· The RNA workflow compensates for DNA workflow limitations by identifying fusion partners and validating functional impacts, ensuring high-confidence variant detection.
Flexible Customization
· Supports subdivision into sub-panels or addition of custom targets to address diverse research and clinical needs.
Gene List
Gene List
CDCap™ Hema-Tumor Expanded Gene Panel_DNA_436 genes_Full CDS
| ABL1 | ABL2 | ACTB | AKT1 | AKT2 | AKT3 | ALK | AMER1 | ANKRD35 | APBB1 | APC | AR | ARAF | ARHGAP26 | ARID2A |
| ARID2B | ARID3 | ARID5B | ASXL1 | ASXL2 | ASXL3 | ATG5 | ATM | ATR | ATRX | ATXN2 | AURKA | AURKB | AURM1 | AXL |
| BLM | BRAF | BRAF1 | BRAF6 | BLC10 | BLC11 | BLC11B | BLC16 | BLC6 | BLC7A1 | BCOR | BCORL1 | BKLC | BLM | BLNK |
| BRPF1 | BRCA1 | BRCA2 | BRD1 | BRD2 | BRD4 | BRIP1 | BUB1B | BUBR1 | CALR | CAROL1 | CCNB1 | CCND1 | CCND2 |
| CCND3 | CCNE1 | CCNE2 | CCNT1 | CCT8B | CD2 | CDT1 | CDT4 | CDK8 | CDK8S | CDK10 | CDK19 | CDK19B | CDK8 | CORC4 |
| COG73 | COPH1 | COPH2 | COPH4 | COPS8 | COPB1 | COPB2 | COPS10 | COPS2 | CONDCA | COND8 | COND28 | COND22 | CEBPA | CEP57 | CHD2 |
| CHD8 | CHEK1 | CHEK2 | CKTA | CKS1B | CNRBP | CRLF2 | CSP1R | CSP3R | CTCF | CTLA4 | CTNNB1 | CUX1 | CCNC | CYLD |
| DAXX | DDR2 | DDX1X | DDX41 | DHX35 | DNAH1 | DMT1A | DOT1L | DTX1 | DUSP2 | DUSP22 | EBF1 | ECT2L | ELO | EGFR |
| EP300 | EPCAM | EPH4B | EPH4A | EPHAS1 | EPHX1 | EMR2 | EMR3 | EMR4 | EMR5 | EMX1 | EMX2 | EMX3 | ESAM | ETV6 | ETSI |
| ETV5 | ETV6 | ETV5 | FAMAC | FANCA | FANCC | FANCD2L | FANCE | FANCF | FANCG | FANCI | FANCL | FANCM |
| FAS | FAT11 | FAT14 | FBXO11 | FBXO7 | FGFR1 | FGFR2 | FGFR3 | FGFR4 | FH | FLCN | FLT1 | FLT3 | FLT4 | FOXO3 |
| FOXD3 | FOXP1 | FN | GA949 | GATA1 | GATA2 | GATA3 | GNAI1 | GNA13 | GNAQ | GNAS | GNB1 | GNB3A | HACE1 | HBA1 |
| HRB2 | HHRP | HNRNPC | HNRNPA2B1 | HIST1H1C | HIST1H1D | HIST1H1E | HNF1A | HRAS | ID3 | IDH1 | IDH2 |
| INHBA | IGF1R | IGH | IKK | IKL | IGL5 | IRF6 | IKZF1 | IKZF2 | IKZF3 | IL3 | IL7R | INPP8B | INPP5D | IRF1 |
| IRF4 | IRF8 | ITK | ITPRB | JAK1 | JAK2 | JAK3 | JARID2 | JUN | KAT6A | KDM2B | KDM5A | KDM6C | KDM6A | KOR |
| KIT | KIT2 | KITL5 | KITL6 | KMT2A | KMT2B | KMT2C | KMT2D | KRAS | LEF1 | LMO2 | LPP1B | LTB | LXN | LYST |
| MAP1 | MAP9 | MAL11 | MAP2K1 | MAP2K2 | MAP2K4 | MAP3K1 | MAP3K14 | MAP3K2 | MCL1 | MDM1 | MDM4 | MEL18 | MZF2B | MZF2D |
| MEN1 | MET | MPHAS1 | MGA | MTF2 | MLH1 | MPL | MSH2 | MSH3 | MSH6 | MTOR | MUTYH | MYB | MYC | MYCL |
| MYCN | MYOD1 | NHSL1 | NCKAP1L | NCSTN | NF1 | NF2 | NFETL3 | NPHS1 | NPHS2 | NPHS4 | NPHVE | NOV-1 | NR4A | NOTCH1 |
| NOTCH2 | NPHS1 | NRAS | NSG1 | NTSC2 | NTRK1 | NTRK3 | NUP98 | PBX8 | PAG1 | PAX5 | PALB2 | PAX5 | PBRM1 | PC |
| PCCD1 | PCCD1LG2 | PDGFRA | PDGFRB | PKD1 | PHF6 | PKDCA | PKDCC | PKLR1 | PKLR2 | PNI1 | PLCG1 | PLCG2 | PML | PHZ2 |
| POLE | POT1 | POLGAM1 | PPM1D | PPARD2A | PRDM1 | PRF1 | PRKCB | PRKPR | PTPN1 | PTPN11 | PTPN6 | PTPN6 |
| PTPRD | PTPRK | PTPRO | RAD51A | RAD21 | RAD50 | RAD51 | RAF1 | RARA | RASEF2A | RB1 | RECQL4 | REL | RELN | RET |
| RHOA | RICTOR | RNF43 | RGS1 | RPL10 | RPL5A | RPTOR | RUNX1 | SAMD9 | SAMD9L | SBDS | SDHA | SDHB | SDHC | SDHD |
| SETBP1 | SETD2 | SF3B1 | SOX1 | SHC2B | SH2D1A | SLIC3H1 | SLCTA1 | SMAD2 | SMAD4 | SMARC2 | SMARC4 | SMARCB1 | SNCA | SNC3 |
| SMG2 | SOCS1 | SOD1L1 | SOD2 | SPEN | SPOP | SRC | SRY12 | SRF2 | STAT2 | STAT3 | STAT4 | STAT5A | STAT5B | STAT6 |
| STK11 | STAT1 | STKBP2 | SULT1 | SUZ12 | SYK | TRIM24 | TGCT | TCL3 | TENC1 | TENT1 | TENT2 | TENT3 | TGFR2 |
| TNKS1 | TNKS1F2 | TNKSNF2 | TNKS1RBF2 | TCPA | TCP4 | TP53 | TP53BP1 | TP73 | TMX4 | TMX5 | TSC1 | TSC2 |
| TSHR | TRX2 | ULK1F | UNC13D | VAV1 | VHL | WN5C1 | WTI | XMP | XPC | XPO1 | ZAP70 | ZBTB3A | ZEB2 | ZMYM3 |
| ZRSR2 |
| ALK | BLC6 | BCR | BRPF1 | ETV6 | JAK2 | KMT2A | MYC | PDGFRA | PDGFRB | RARA | RET |
| ABL1 | ABL2 | ALK | ARHGAP26 | ARID1A | ASXL1 | ATG5 | BCL10 | BCL11A | BCL11B | BCL2 | BCL3 | BCL6 | BCL7A | BCL9 |
| BCOR | BCR | BIRC3 | BRAF | BTG1 | CBFA2T3 | CBFB | CBL | CCND1 | CCND2 | CCND3 | CD274 | CDK6 | CEBPE | CITTA |
| CREBBP | CRLF2 | CTNNB1 | DEK | DUSP22 | DUX4 | EGFR | EP300 | EPOR | ERBB2 | ERG | ETS1 | ETV1 | ETV4 | ETV6 |
| EWSR1 | FGFR1 | FGFR2 | FGFR3 | FOXO1 | FOXO3 | FOXP1 | GLIS2 | HLF | IGH | IGK | IGL | IKZF1 | IL3 | IRF4 |
| ITK | JAK1 | JAK2 | JAK3 | KAT6A | KIF5B | KMT2A | LMO1 | LMO2 | LYN | MAF | MAFB | MALT1 | MECOM | MEF2D |
| MLLT1 | MLLT10 | MLLT3 | MN | MTCF1 | MYB | MYC | MYH11 | NCOA2 | NCOR1 | NF1 | NF2 | NFKB2 | NOTCH1 | NPM1 |
| NSD1 | NTRK1 | NTRK3 | NUP214 | NUP98 | NUTM1 | P2RY8 | PAX5 | PBX1 | PDCD1LG2 | PDGFRA | PDGFRB | PICALM | PIM1 | PML |
| POU2AF1 | PRDM1 | PTCH1 | RAF1 | RARA | RARB | RARG | RBM15 | RET | ROS1 | RUNX1 | RUNX1T1 | STAT6 | STIL | SYK |
| TAF15 | TAL1 | TBL1XR1 | TCF3 | TCL1A | TCL1B | TET1 | TLX1 | TLX3 | TNFRSF11A | TOP1 | TP63 | TPM3 | TPM4 | TRA |
| TRB | TRD | TRG | TYK2 | VAV1 | WHSC1 | ZBTB16 | ZEB2 | ZNF292 | ZNF362 | ZNF384 |
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.