Custom Macular Degeneration Panel
Introduction
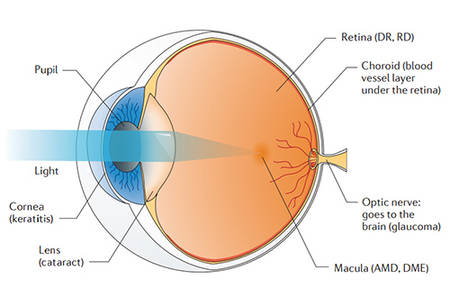
The macular area is a normal physiological area on the retina of the fundus. It is located in the center of the retina and concentrates on a large number of visual functional cells. Macular degeneration is the degeneration of the central region of the retina, which can eventually lead to visual impairment, or even a certain chance of blindness. The macular disease can be caused by hereditary lesions, senile changes, inflammatory lesions and other fundus lesions. Hereditary lesion can show symptoms from early childhood to old age, but the most common is adolescent-onset, and it is hard to cure. Age-related Macular Degeneration (AMD) can be ameliorated or stabilized through early diagnosis and appropriate treatment. Inflammatory macular lesions are more common in various retinal choroiditis, such as toxoplasmosis, uveitis, etc. In addition, retinal vasculitis, visual venous occlusion, diabetic retinopathy, high myopia and traumatic rupture of the choroid can also cause damage in the macular area. Macular degeneration is divided into two types: "dry" and "wet". The two types of lesions are related to the coordination between multiple genes, and also related to environmental factors. Symptoms associated with the early stage of the lesion are sudden loss of vision and visual impairment. Because there is no particularly effective cure for macular degeneration, earlier determination is more conducive to slowing the progression of the disease.
Disease-related gene description
Genetic analysis revealed that five genetic mutations on chromosomes 1, 6, and 10 increased the risk of macular degeneration by at least 50%. These genes are involved in the regulation of immune responses, the inflammatory response and the maintenance of retinal homeostasis. Mutants with different dysfunctions accumulate over time, causing accumulation of metabolic wastes which lead to retinal damage. For example, the complement factor H gene (CFH) located on chromosome 1 of 1q31.3. The common mutation of this gene is closely related to the production of AMD. Mutations in CFH (Y402H) results in decreased ability of CFH to regulate complement on retinal surface, which leads to increased macular inflammation. Dry macular degeneration is associated with the death of visual cells. The clinical manifestation of wet macular degeneration is fundus neovascularization. Studies have shown that pathological activity is related to mutations in visual cell nutrition, energy supply, excessive free radical production, stress and other related genes, such as mutations in CDH3, DRAM2, IMPG1, IMPG2, PRDM13, PRPH2, RAX2, RLBP1, and TIMP3 which can lead to an increased disease risk. In addition, genes such as ABCA4, CERKL, CRB1, IMPG2, PROM1, LIPC, PRPH2, RDH12, RP1L1 and RPGR involved in the regulation of expression of pigments, ion channels, lipid kinases, can also cause lesions. Detecting these mutations can help to predict the trend of the lesion.
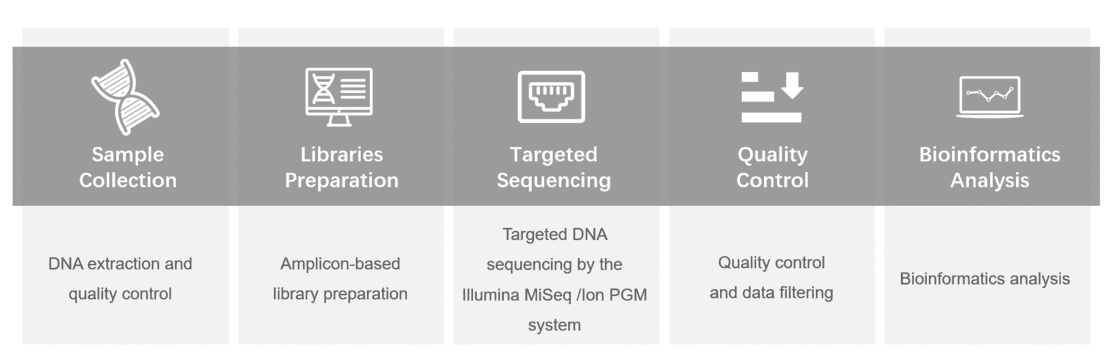
To better understand the genetic variants of macular degeneration associated genes, you can choose genes from the macular degeneration panel content below for detecting. Our authoritative team provides targeted DNA sequencing and detailed analysis reports, using the Illumina MiSeq or Ion PGM system.
Custom macular degeneration panel offers but are not limited to:
-
Superior analytical performance includes advanced enrichment methods, powerful sequencing technology and precise bioinformatics pipelines.
-
Cover multiple mutations and have the capability to detect low frequency of mutational variants consistently with requirement.
-
Every detected genetic variant is selected from authoritative clinical mutation data and current scientific researches.
-
Contact details of test performance are publicly available.
Choose the genes that suit you from the macular degeneration gene list
| ABCA4 |
BEST1 |
CRX |
CTNNA1 |
CDH3 |
| C1QTNF5 |
CERKL |
CNGB3 |
CRB1 |
CRX |
| CTNNA1 |
DRAM2 |
CFB |
CFH |
EFEMP1 |
| ELOVL4 |
FBLN5 |
PRDM13 |
HTRA1 |
IMPG1 |
| IMPG2 |
LIPC |
LRP6 |
MFSD8 |
MT-ND2 |
| PARP12 |
PROM |
PRPH2 |
RAX2 |
RCA |
| RDH12 |
RHD5 |
RLBP1 |
RPGR |
RS1 |
| TIMP3 |
VEGF |
VLDLR |
|
|
Specimen requirements of our custom macular degeneration panel
- Specimen: blood, saliva or extracted DNA.
- Volume: 3 mL blood, 2 mL saliva and 3ug DNA.
- Collection: blood is collected by routine blood collection and saliva is collected by saliva collection kits (kits are available upon request). DNA samples are stored in TE buffer or equivalent.
- Container: lavender-top (EDTA) tube or yellow-top (ACD) tube.
- Storage/transport temperature: room temperature.
Gene panel workflow
For more information about the Custom Macular Degeneration Panel or need other amplification requirements, please contact us.
References:
- Fine A M, Elman M J, et al. Earliest symptoms caused by neovascular membranes in the macula. Archives of ophthalmology, 1986, 104(4): 513-514.
- Hughes A E, Orr N, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nature genetics, 2006, 38(10): 1173.
- Yates J R W, Sepp T, et al. Complement C3 variant and the risk of age-related macular degeneration. New England Journal of Medicine, 2007, 357(6): 553-561.
- Yang Z, Camp N J, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science, 2006, 314(5801): 992-993.
- Hirschler B. Gene discovery may help hunt for blindness cure. Reuters. 2008: 10-07.
- Udar N, Atilano S R, et al. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Investigative ophthalmology & visual science, 2009, 50(6): 2966-2974.
- Allikmets R, Shroyer N F, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science, 1997, 277(5333): 1805-1807.
- Sprecher E, Bergman R, et al. Hypotrichosis with juvenile macular dystrophy is caused by a mutation in CDH3, encoding P-cadherin. Nature genetics, 2001, 29(2): 134.
- Ayyagari R, Demirci F Y, et al. X-linked recessive atrophic macular degeneration from RPGR mutation. Genomics, 2002, 80(2): 166-171.
- Sidjanin D J, Lowe J K, et al. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Human molecular genetics, 2002, 11(16): 1823-1833.
- Kuehn M H, Stone E M, et al. Organization of the human IMPG2 gene and its evaluation as a candidate gene in age-related macular degeneration and other retinal degenerative disorders. Investigative ophthalmology & visual science, 2001, 42(13): 3123-3129.
- Weber B H F, Vogt G, et al. Sorsby's fundus dystrophy is genetically linked to chromosome 22q13–qter. Nature genetics, 1994, 7(2): 158.
- Haines J L, Schnetz-Boutaud N, et al. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Investigative ophthalmology & visual science, 2006, 47(1): 329-335.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources