Amplicon Sequencing-Based NGS Panel
CD-Genomics is a company with rich experience in targeted gene panel sequencing which is used to accurately detect gene variations related to diseases.
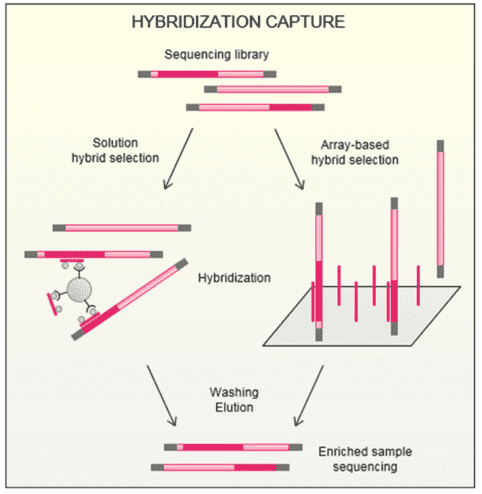
CD-Genomics provides custom NGS panels for mutations in at least 68 diseases and thousands of genes. We can employ two target enrichment strategies prior to the NGS panel, which are amplification and hybridization capture. Embarked with great primer-probe specificity, high on-target sequencing reads can be obtained by amplicon-based enrichment, which has been preferentially used on the basis of short preparation time and small DNA input amounts. We can offer fast, accurate, and cost-effective detection of disease-related genes of your interest based on amplicon sequencing strategy.
In an amplicon-based target enrichment assay, genomic DNA is firstly fragmented by enzymatic or mechanical methods. The following step involves the amplification of the regions of interest using sequence-specific primers, where regions of interest could vary greatly among exome, rRNA, miRNA, etc. In this step, PCR runs in single or multiplex reactions, generating single or multiple amplicons. Then amplicons representing each region of interest are pooled to create a library of fragments for sequencing.
Highlight Applications
For smaller gene panels or mutation hotspots
Genotyping by sequencing
Detecting disease-associated variants
Detecting germline inherited SNPs and indels
Amplicon Sequencing-Based Disease Panel Workflow

Amplicon sequencing is used for genotyping by sequencing and for the detection of germline SNPs, indels, and known fusions. It is particularly suited to the detection of disease-associated variants. Our strict quality control throughout the pipeline workflow to ensure the accuracy and repeatability of the enrichment, sequencing and bioinformatics analysis of amplicon sequencing-based NGS panel.
Features of Our Custom Amplicon Sequencing Panel
Highly accurate amplicon sequencing panel for scientific research purposes
Low DNA input and high on‐target rates
Easily accessible / Effective in enrichment and specificity
Ultra-deep sequencing of target specific genomic regions
Sample Requirements
- DNA > 50 ng, no obvious degradation, 1.8 < OD260/280 < 2.0, OD260/230 ≥ 1.8, concentration ≥ 30 ng/μL.
- Or properly treated blood or tissue samples.
Bioinformatics Analysis
| Analysis Contents |
Details |
| Probe design |
The design of probes with complementary sequence to the flanking regions of the target DNA to be sequenced. |
| Primary analysis |
To identify single nucleotide variants and small insertions/deletions. |
| Trimming pipeline |
Quality control check is implemented and only samples with enough reads and base quality are considered |
| Variant identification & annotation |
The alignment of each read with the expected amplicon. Mutations are considered pathogenic if they are absent from controls, predicted to alter the sequence of the encoded protein and to adversely affect protein function. |
| Allelic variant profiling |
To detect and characterize complex allelic variants, such as short tandem repeats. |
| Variant classification |
To detect mutations at low-allelic fraction from amplicon sequencing of matched tumor-normal sample pairs. |
Why choose us?
- Advanced technology platform and experienced team of scientists
- Customized services for the detection of diverse disease genes
- Bioinformaticians can get you more valuable information from the NGS panel
- Affordable price and fast turnaround time
Select and submit the target genes you are interested in and we will send you a design coverage report and provide you with a quote. If you have any questions or requirements, please feel free to contact us, we will help you solve the problems and further optimize to meet your needs.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources