Custom Spinocerebellar Ataxia Panel
What is spinocerebellar ataxia?
Spinocerebellar ataxia (SCA) is a genetic disorder characterized by slow progressive gait. It is often associated with poor coordination of hand, speech and eye movements. Spinocerebellar ataxia causes great inconvenience to the life of patients and their families. Spinocerebellar ataxia has many types, and each of them could be considered a neurological condition in its own right. This disease is caused by the mutation of either a recessive or dominant gene. SCA can affect anyone of any age, and is currently considered to be a progressive and irreversible disease. There is no cure for SCA till now. The treatment now we use are directly towards alleviating symptoms, but can’t cure the spinocerebellar ataxia. The study of spinocerebellar ataxia-related genes is an important way to help us explore the pathogenesis of SCA and develop new therapies.
Disease-related gene description
Although there are many types of SCA, there are not many related genes. Till now we find 5 genes related, they are ATXN1, ATXN2, ATXN3, ATXN6, ATXN7, respectively. The different loss-of-function mutations in ATXN1 have distinct phenotypes, which explains why researchers only find five strong-related genes but SCA has many types. Protein encoded by ATXN1 forms a transcriptional repressor complex with Capicua, and the complex leads to neurodegeneration which makes SCA happens. The mutations of CAG repeat expansions in the middle sequence of ATXN2 and ATXN3 can also increase the risk of SCA. Most of the mutations in SCA-related genes cause the repeat expansions of trinucleotide. These expansions occur between two generations of unaffected individuals and promote the emergence of an ataxia phenotype. But ATXN7 appears to cause SCA in a different way. Mutations in ATXN7 cause the expansion of the polyglutamine tract and lead to the development of debilitating neurodegenerative diseases.
CD-Genomics provides a custom SCA panel which contains optimized genes related to SCA. You can customize the gene panel depend on your own requirements. Targeted sequencing technology powered by Illumina MiSeq system/Ion PGM system ensures to investigate the SCA related genetic variations discovered in routine clinical practice.
Custom SCA Panel offers but are not limited to:
-
Automation and high-throughput amplicon sequencing by Illumina MiSeq system/Ion PGM system is provided.
-
Only sequencing the customizable SCA panel meets your requirements, increases throughput and saves costs.
-
Strict quality control throughout the pipeline workflow ensures the accuracy and repeatability of the sequencing.
-
Every detected genetic variant will be further validated to ensure the validity of results.
-
Custom panel content is designed to keep up with the frontiers from current literature about SCA to target all relevant regions.
-
You can choose the panel content from our SCA library or discuss your custom SCA requirements, then we can provide you with your own panel.
Choose the genes that suit you from the SCA gene list
| ATXN1 |
ATXN2 |
| ATXN3 |
ATXN6 |
| ATXN7 |
|
Specimen requirements of our custom SCA panel
- Specimen: blood, saliva or extracted DNA (we do not accept DNA samples isolated from FFPE tissue).
- Volume: 2-5 mL blood, 2 mL saliva, 3ug DNA.
- Collection: blood is collected by routine blood collection and saliva is collected by saliva collection kits (kits are available upon request). DNA samples are stored in TE buffer or equivalent.
- Container: lavender-top (EDTA) tube or yellow-top (ACD) tube.
- Storage/transport temperature: room temperature.
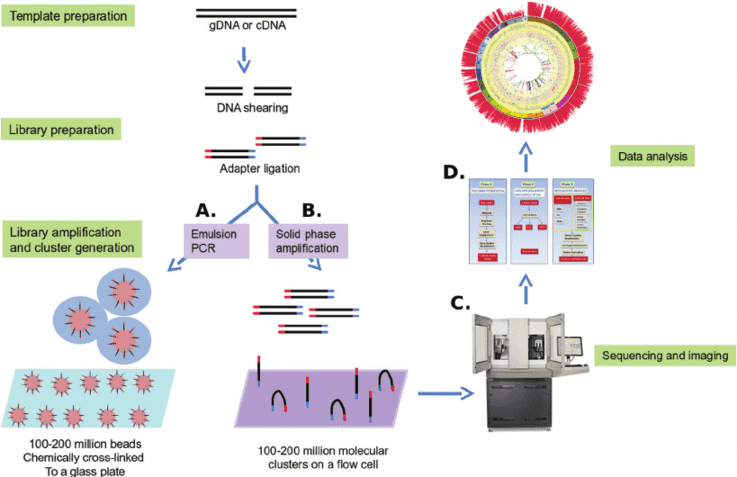
Gene panel workflow
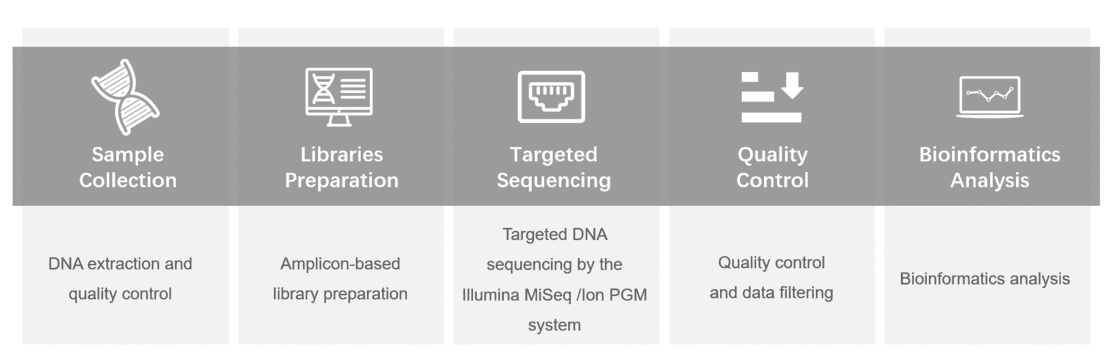
For more information about the Custom SCA Panel or need other amplification requirements, please contact us.
References:
- Lu HC, et al. Disruption of the ATXN1–CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nature Genetics, 2017, 49(4):527-536.
- Van Damme P, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology, 2011, 76(24):2066-2072.
- Stochmanski S J, et al. Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Human Molecular Genetics, 2012, 21(10):2211.
- Eldamária de Vargas Wolfgramm, et al. Molecular analysis of spinocerebellar ataxia trinucleotide repeat behavior in normal individuals of a Brazilian population. Journal of the Neurological Sciences, 2008, 269(1-2):113-117.
- Lan X, et al. Poly(Q) Expansions in ATXN7 Affect Solubility but Not Activity of the SAGA Deubiquitinating Module. Molecular and Cellular Biology, 2015, 35(10):1777-1787.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources