Custom Hearing Loss Research Panel

What is hearing loss?
Hearing loss (HL) is the most common sensory defected disease and currently disables 466 million people across the world. This illness also affects newborns severely. Approximately 1-3 babies are persecuted in every 1000 newborns worldwide. There are lots of reasons for HL, such as Cranio-facial abnormalities and congenital infections, but half of HL patients are attributed to genetic factors. HL disease could be divided into the syndromic and the non-syndromic according to the accompanying signs and symptoms. Non-syndromic hearing loss (NSHL) accounts for a large proportion of HL. NSHL is caused by multiple pathogenic factors. It broadly separates into different modes of inheritance, including autosomal recessive (AR), autosomal dominant (AD), X-linked and mitochondrial. The mutations of hearing loss-related genes will lead to protein inactivation and abnormal signaling pathways, thus resulting in the only clinical symptom: hearing loss.
Disease-related gene description
It's reported that non-syndromic hearing loss has no malformations of the external ear or any related medical problems, however, it may be associated with abnormalities of the middle ear and/or inner ear. NSHL has four typical modes of inheritance. The inheritance pattern among the disorders with prelingual non-syndromic hearing loss is 80%. ACTG1, CD164, GJB2 KCNQ4, and WFS1 are related to autosomal dominant non-syndromic hearing impairment, most of which can cause post-lingual hearing impairments. Autosomal recessive non-syndromic deafness also has extreme genetic heterogeneity. GJB2 is one of the typical genes. It possesses high-frequency of pathogenic variants. PRPS1, POU3F4, SMPX, AIFM1 and COL4A6 are X-linked non-syndromic hearing impairment-related genes. They can be either pre- or post-lingual. Most of the pathogenic variants in mitochondrial genes result in an extensive spectrum of maternally inherited multisystem disorders. However, mitochondrial non-syndromic hearing impairment is caused by the mutations in MT-RNR1, MT-TS1 and MT-CO1. The MT-CO1 mutations present extremely severe symptoms. The other two genes have many mutations of varying severity.
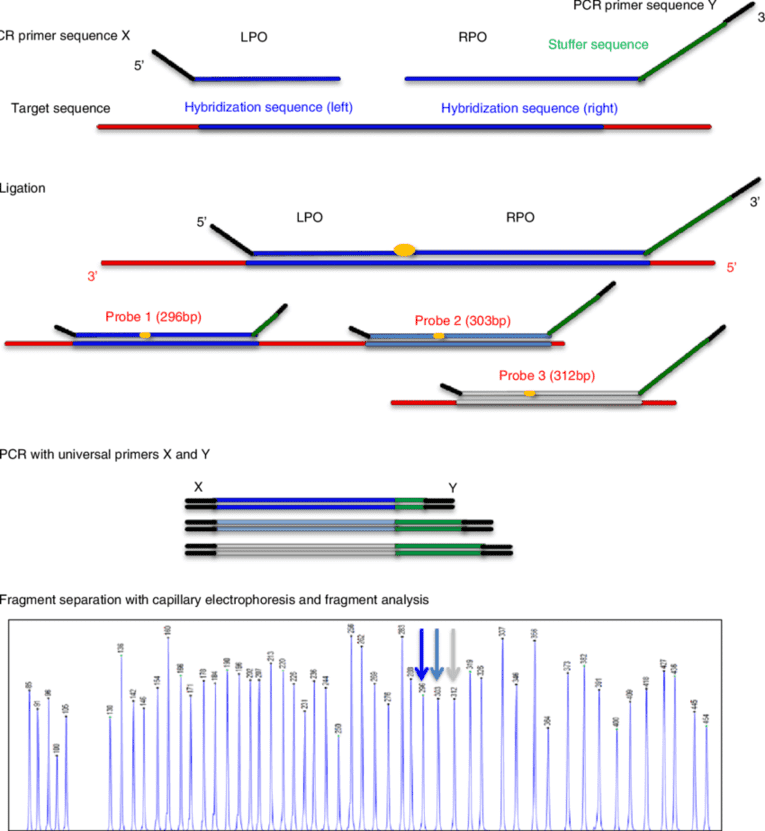
Hearing loss related genes are abundant and their mutations are complicated. Targeted sequencing will help you accelerate your biology researches on hearing loss. Our platform provides targeted DNA sequencing by the Illumina MiSeq, and offers an abundant hearing loss research panel library. You can choose the genes you are interested in to detect the genetic variation information.
Custom hearing loss research panel offers but are not limited to:
-
Provide gene content related information consultation and customize panel service, increases throughput and saves costs.
-
Utilize the unparalleled coverage uniformity of targeted enrichment DNA sequencing by the Illumina MiSeq to provide a highly targeted sequencing, which is developed as a quick, accurate and cost-effective method to identify genetic mutations associated with hearing loss.
-
To make sure the accuracy and repeatability of the sequencing, we will take strict control throughout the pipeline workflow.
-
The results of detected genetic variant will be further validated to verify the sequencing accuracy.
-
Custom your hearing loss research panel in line with the frontiers from current literature and keep in step with the latest research results to ensure the innovation of your panel.
Choose the genes that suit you from the hearing loss research gene list
| ACTG1 |
ADGRV1 |
BTD |
CCDC50 |
CD164 |
| CDH23 |
CEACAM16 |
CEP78 |
CHD7 |
CIB2 |
| CISD2 |
CLDN14 |
CEACAM16 |
CIB2 |
CLRN1 |
| COCH |
COL11A2 |
DIAPH1 |
DMXL2 |
DSPP |
| EDN3 |
EDNRB |
EYA1 |
EYA4 |
GJB2 |
| GJB3 |
GJB6 |
GRHL2 |
GSDME |
HARS1 |
| HOMER2 |
KCNE1 |
KCNQ1 |
KCNQ4 |
MIR96 |
| MITF |
MYO7A |
NF2 |
PAX3 |
PCDH15 |
| PEX7 |
PHYH |
SIX1 |
SIX5 |
|
(Download the hearing loss gene list for more genes)
Specimen requirements of our custom hearing loss research panel
- Specimen: blood, DNA or tissue.
- Collection: 3-5 mL draw blood, 5 µg genomic DNA or 50 mg tissue for DNA analysis.
- Container: EDTA (purple-top) tube(s) for blood and DNA samples, tissues should be flash frozen in liquid nitrogen at collection.
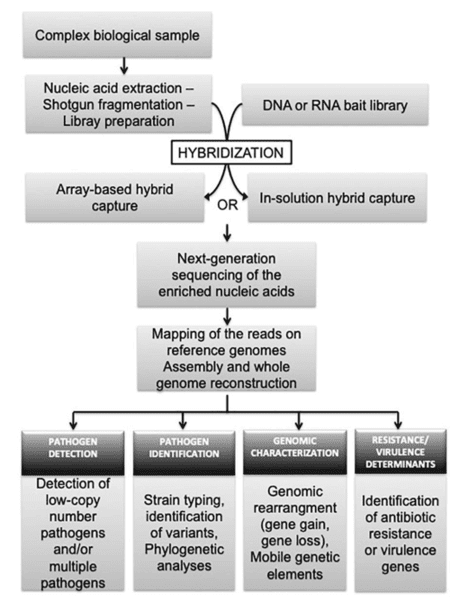
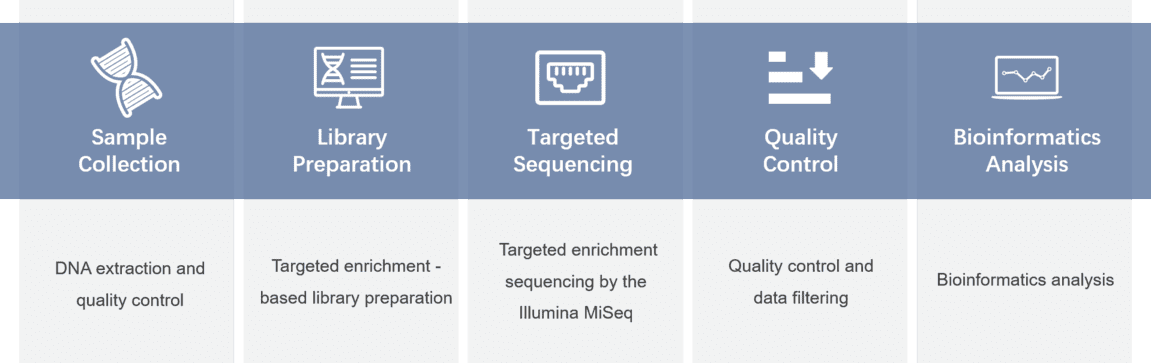
Gene panel workflow
For more information about the Custom Hearing Loss Panel or need other amplification requirements, please contact us.
References:
- Resendes BL, et al. At the speed of sound: gene discovery in the auditory system. American Journal of Human Genetics, 2001, 69(5):923-35.
- Dunbar LA, et al. Clarin-2 is essential for hearing by maintaining stereocilia integrity and function. Embo Molecular Medicine, 2019.
- Korver AM, et al. Congenital hearing loss. Nature Reviews Disease Primers, 2017, 3:16094.
- Ren Y, et al. Gene Therapy for Human Sensorineural Hearing Loss. Frontiers in Cellular Neuroscience, 2019, 13:323.
- Sun W, et al. Hearing loss and auditory plasticity. Hear Res, 2017, 347:1-2.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources