Deciphering the Role of Long Non-Coding RNA in Cancer Research
Long non-coding RNAs, abbreviated as long ncRNA or lncRNA, are known as transcripts composed of more than 200 nucleotides and are not translated into protein. Having more than 200 nucleotides, lncRNAs are distinguished from short or small non-coding RNAs such as small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), and small nucleolar RNAs (snoRNAs).
Long ncRNAs are regarded to be more nuclear-localized as compared to messenger RNAs (mRNAs) which is the reason for inefficient splicing and polyadenylation, as well as susceptibility to derogation, specifically, by exosomes on chromatin. Aside from that lncRNAs usually have complex secondary and tertiary structures, while their functions only impose very subtle sequence constraints. There are several ways to detect lncRNA, either through chemical or enzymatic probing. Its detection via probing is very crucial as it provides an understanding of the secondary structure. One of the most recent methods used in sequencing long ncRNAs is through next-generation sequencing approaches, such as amplicon sequencing.
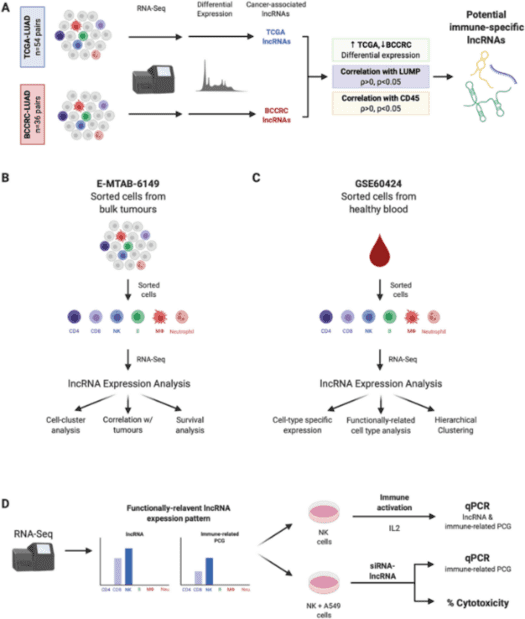 Figure 1. Overall analysis process of long non-coding RNA expression revealing novel mediators of the lung tumour immune response. (Sage, 2020)
Figure 1. Overall analysis process of long non-coding RNA expression revealing novel mediators of the lung tumour immune response. (Sage, 2020)
Functions of Long Non-Coding RNAs
Long ncRNAs, especially those with unconventional forms since they accumulate in the nucleus, are emerging as crucial regulators in gene expression networks as it controls nuclear architecture and transcription in the nucleus, as well as it modulates inter-and intrachromosomal interactions such as mRNA stability, translation, and post-translational modifications in the cytoplasm. These lncRNAs also have toles in chromatin modeling. Specifically, they promote the chromatin modifiers and prevent the recruitment of chromatin modifiers. In terms of translation regulation, lncRNAs play an important role in regulating transcription by forming R-loops, interfering with Pol II machinery, as well as in transcription or the lncRNA locus regulate transcription. LncRNAs also have roles in the regulation of nuclear bodies. These include acting as architectural RNAs, as well as acting as non-architectural lncRNAs in nuclear bodies. Other functions of lncRNAs in the cytoplasm include regulating mRNA turnover, interfering with posttranslational modifications (PTMs), and regulating translation.
Association of Long Non-Coding RNA in Cancer
A lot of long ncRNAs have been functionally associated with human diseases, especially with cancer. Up to date, the dysregulation of long ncRNAs has been implicated with several types of cancer: (1) glioblastoma, (2) colorectal cancer, (3) ovarian cancer, (4) breast cancer, (5) lung cancer, and many other types. The dysregulation of long RNAs was known to have impacts on cellular functions. These include cell proliferation, induction of angiogenesis, promotion of metastasis, resistance to apoptosis, and evasion of tumor suppressors.
Long ncRNAs work through multiple mechanisms, particularly in cancer. These include, but not limited to, chromatin remodeling, completing endogenous RNAs (ceRNAs), natural antisense transcripts (NATs), and chromatin interactions. As mentioned, long ncRNAs can interfere with the chromatin remodeling machinery in many ways. These include acting as signal long ncRNAs or scaffold long ncRNAs. Another mechanism action by which long ncRNAs function to regulate transcription has been recently discovered. The said function is to stabilize, as well as maintain chromatin loops as it involves the production of long ncRNAs from the eRNAs or the enhancer regions in the genome. The forming of chromatin loops enables distally located enhancers, which some are located hundreds of kilobases away, to interact with their target gene promoters. One way to target specific genes or mutations which are associated with cancer phenotype is by using cancer panels that utilize next-generation sequencing technology.
The profiles of long ncRNAs can also be used in identifying clinically relevant cancer subtypes, particularly those that can predict tumor behavior and disease prognosis. A good example would be the overexpression of the long ncRNA SChLAP1 in men treated with radical prostatectomy. This overexpression was associated with a 2.45-fold increased chance of developing metastatic progression which is similar to the risk associated with high-grade prostate cancer.
References:
- Sage AP, Ng KW, Marshall EA, et al. Assessment of long non-coding RNA expression reveals novel mediators of the lung tumour immune response. Scientific reports. 2020, 10(1):1-3.
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nature cell biology. 2019, 21(5).
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer cell. 2016, 29(4).
- Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013, 193(3).
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
 Figure 1. Overall analysis process of long non-coding RNA expression revealing novel mediators of the lung tumour immune response. (Sage, 2020)
Figure 1. Overall analysis process of long non-coding RNA expression revealing novel mediators of the lung tumour immune response. (Sage, 2020)