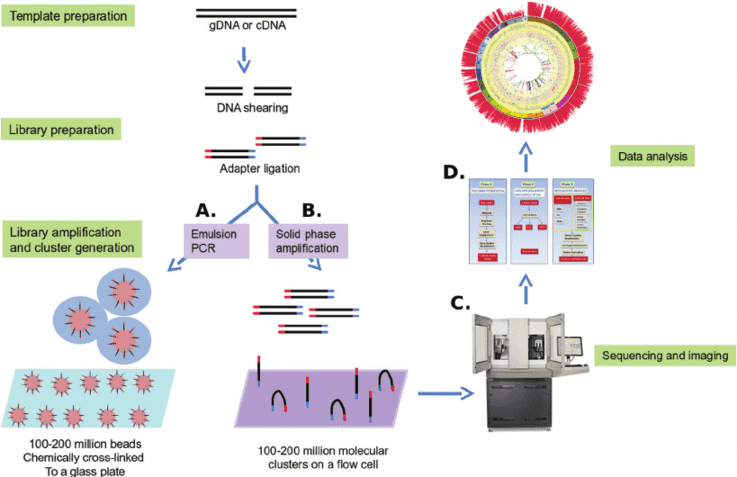
We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Navigation
- Home
- Products
- Ready-to-Use NGS Panel
- Real-Time PCR Kits
- Discover the infinite power of our Respiratory Virus Kits to drive scientific discovery
- Cancer Real-Time PCR Kits
- Pathogens Detection
- Respiratory Pathogen RT-PCR Kits
- Respiratory Virus RT-PCR Kits
- EBV RT-PCR Kits
- HBV RT-PCR Kits
- HPV RT-PCR Kits
- HLA Typing Kits
- Food & Environmental Microbiology Test Kits
- Pharmacogenomics Testing Kits
- Allergens Detection Kits
- Food & Animal Feed Ingredients Authentication Kits
- Plant GMO Detection Kits
- Oncology Quality Control Products
- Pathogenic Microorganisms Control Products
- QC & RM for Oncology Assays
- Copy Number Variation (CNV) Reference Products
- ctDNA (Circulating Tumor DNA) Reference Products
- Tumor Companion Diagnostic Reference Products
- Homologous Recombination Deficiency (HRD) Reference Products
- Minimal Residual Disease (MRD) Reference Products
- Homologous Recombination Repair (HRR) Reference Products
- Tumor Mutation Burden (TMB) Reference Products
- Microsatellite Instability (MSI) Reference Products
- Reproductive Health Reference Products
- Infectious Diseases Reference Products
- Inherited Diseases Reference Products
- Services
- Predesigned NGS Panel
- Inherited Disease Panel
- Discovery Cancer Panels
- Pan-Cancer Panel Sequencing
- Hereditary Cancer Panel Sequencing
- Cancer Hotspot Panel Sequencing
- Lung Cancer Panel Sequencing
- Breast Cancer Panel Sequencing
- Ovarian Cancer Panel Sequencing
- Thyroid Carcinoma Panel Sequencing
- Esophageal Cancer Panel Sequencing
- Glioma Gene Panel Sequencing
- Colorectal Cancer Panel Sequencing
- Pharmacogenomics Testing
- Pathogens Detection
- Tumor Mutational Burden Analysis
- Solid Tumor Sequencing Service
- Non-NGS Panel
- Custom NGS Panel
- Bioinformatics Analysis Service
- Gene Fusion Detection by Sequencing
- Microsatellite Instability (MSI) Analysis
- Rare Disease Genomics
- SNP Panel & Sequencing
- Predesigned NGS Panel
- Resource
- Company
- Online Inquiry
- Order Online
x