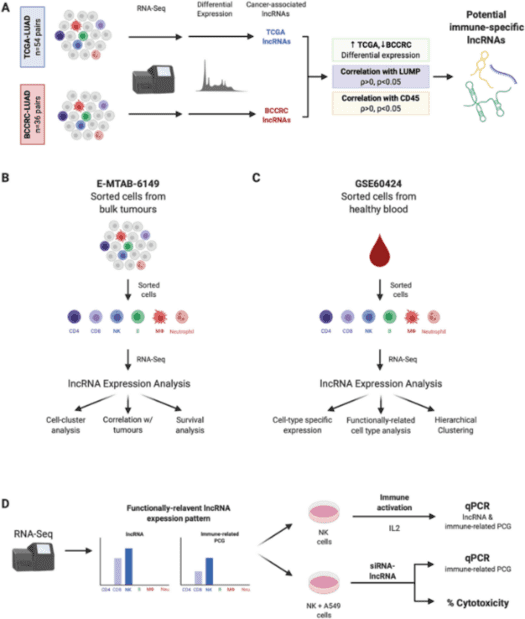
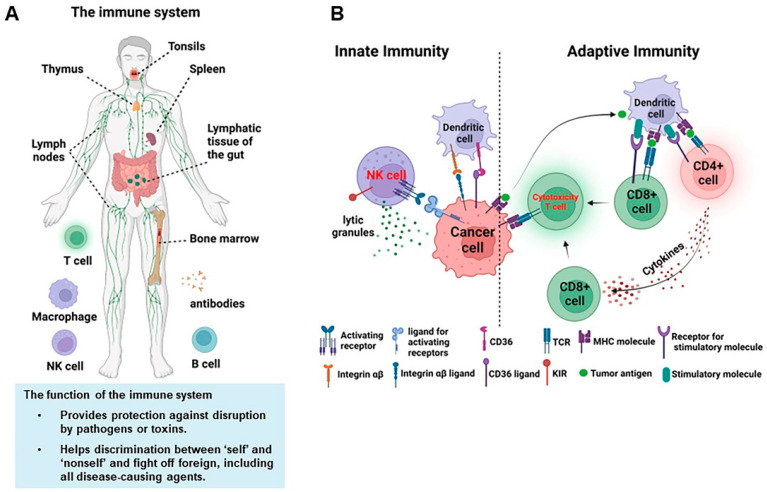
Custom Charcot-Marie-Tooth Panel
What is Charcot-Marie-Tooth?
Charcot-Marie-Tooth, the most frequent hereditary motor and sensory neuropathies disease, is a heterogeneous disorder of the peripheral nervous system. Peripheral nerves could link the brain or spinal cord to muscles or sensory cells, which makes it easy to detect sensations such as touch, pain, heat and sound. Damage to the peripheral nerves worsens over time, which can result in alteration or loss of sensation. According to the electrophysiological and nerve biopsy studies, two major subtypes, demyelinating (CMT1 or HMSN I) and axonal (CMT2 or HMSN II), can be distinguished. CMT1 and CMT2 are usually inherited by an autosomal dominant (AD) trait, but autosomal recessive (AR) and X-linked inheritance have also been reported. More than 40 CMT genes have been identified and several genes associated with related diseases have been identified.
Disease-related gene description
In particular, the disruption of Ca2+ homeostasis has been identified as an important causative factor in the pathophysiology of CMT disease. TRPV4 gene mutations could mediate Ca2+ influx which will result in intracellular toxic effects. PMP22-mediated overexpression of the P2X7 purinoceptor, resulting in high intracellular Ca2+ concentrations in Schwann cells, is a cause of CMT1A. Mutations in the FGD4 gene cause autosomal recessive demyelinating peripheral neuropathy, called CMT4H, which is characterized by its onset and slow progression in infancy or early childhood. LRSAM1 gene encodes for an E3 ubiquitin ligase, which is important for neural function. Inactivation of E3 ubiquitin ligase is characterized by onset in adulthood with slowly progressive length-dependent motor deficit, atrophy, sensory loss to all modalities and foot deformities.
Charcot-Marie-Tooth & Neuropathies are extremely complicated, which makes related gene detection challenging. Next-generation sequencing (NGS) is a strategy that can overcome this problem. Illumina MiSeq's revolutionary process and unparalleled accuracy make it an ideal platform for fast and cost-effective genetic analysis in a wide range of applications. Our platform provides targeted DNA sequencing by the Illumina MiSeq and offers an abundant Charcot-Marie-Tooth & Neuropathies panel library from which you can choose what you want for your researches.
Custom Charcot-Marie-Tooth panel offers but are not limited to:
- Provide related gene information consultation and custom panel services to save your cost and increase efficiency.
- Targeted enrichment sequencing technology and the Illumina MiSeq system can be provided. It is developed as a quick, accurate and cost-effective method to identify genetic mutations associated with Charcot-Marie-Tooth & Neuropathies.
- The results of detected genetic variant will be further validated to verify the sequencing accuracy.
- Custom panel genes are constantly updated based on the frontiers of literature studies to target all relevant regions.
Choose the genes that suit you from the Charcot-Marie-Tooth gene list
| AIFM1 |
ATL1 |
ATL3 |
BSCL2 |
AARS |
| AMACR |
ARHGEF |
ARHGEF10 |
ATAD3A |
ATP7A |
| BAG3 |
BICD2 |
BSCL2 |
C12ORF65 |
CCT5 |
| CHCHD |
COX10 |
COX6A1 |
COX6M |
CTDP1 |
| DCAF8 |
DCTN1 |
DCTN2 |
DHTKD1 |
DNM2 |
| DNMT1 |
DRP2 |
DST |
DYNC1H1 |
EGR2 |
| FAM134B |
FBLN5 |
FGD4 |
FIG4 |
FXN |
| GAN |
GARS |
GDAP1 |
GJB1 |
GNB4 |
| GNE |
GPR49 |
HADHB |
HARS |
HINT1 |
| HK1 |
HNPP |
HSPB1 |
HSPB3 |
HSPB8 |
(Check the Charcot-Marie-Tooth gene list for more genes.)
Specimen requirements of our custom Charcot-Marie-Tooth panel
- specimen: whole blood, saliva or DNA.
- Volume: 3 mL whole blood, 2 mL of saliva and 8 µg of DNA.
- Container: EDTA purple-top or pink-top tubes.
Gene panel workflow
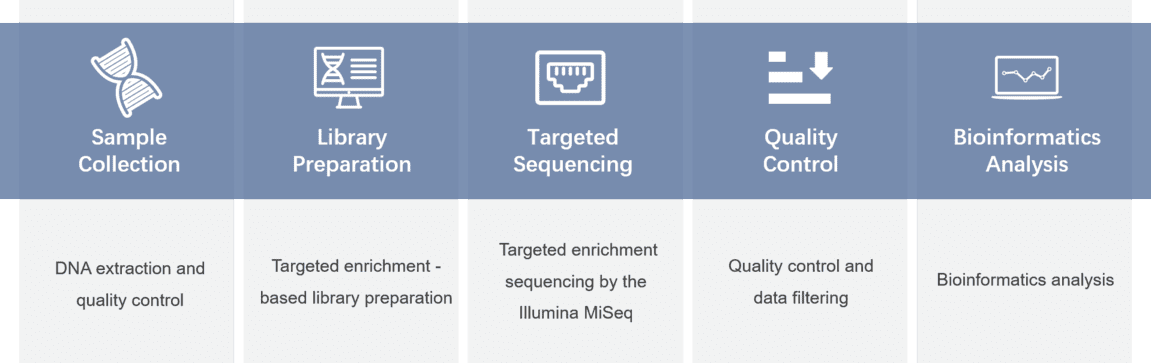
For more information about the Custom Charcot-Marie-Tooth Panel or need other amplification requirements, please contact us.
References:
- Callegari I, et al. Mutation update for myelin protein zero-related neuropathies and the increasing role of variants causing a late-onset phenotype. Journal of neurology, 2019.
- Fernandez-Lizarbe S, et al. Neuroinflammation in the pathogenesis of axonal Charcot-Marie-Tooth disease caused by lack of GDAP1. Experimental Neurology, 2019, 320:113004.
- Sun SC, et al. Mutations in C1orf194, encoding a calcium regulator, cause dominant Charcot-Marie-Tooth disease. Brain, 2019,142(8):2215-2229.
- Previtali SC, et al. Expanding the spectrum of genes responsible for hereditary motor neuropathies. Journal of Neurology Neurosurgery and Psychiatry, 2019, 90(10):1171-1179.
- Peretti A, et al. LRSAM1 variants and founder effect in French families with ataxic form of Charcot-Marie-Tooth type 2. European Journal of Human Genetics, 2019, 27(9):1406-1418.
* For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.
Related Services
Related Products
Related Resources