Whole-Genome SNP Genotyping
Introduction
Single nucleotide polymorphism (SNP) is an important material in genetics research. In recent years, the development of genome-wide SNP marker development methods has enabled researchers to obtain abundant genomic markers at a lower cost, which has greatly promoted related research at the genomic level. Genome prediction establishes training models from individuals with known genotype data and phenotype data, and predicts the genotype and phenotype of individuals with unknown phenotypes, which is of great significance in the field of disease research. Genome-wide genotyping can provide an overview of the entire genome, enabling genome-wide discovery and association studies. Using high-throughput next-generation sequencing (NGS) and microarray technology, researchers can gain a deeper understanding of the genome to explore the functional consequences of genetic variation.
At present, genome-wide association analysis (GWAS) based on microarray has become a commonly used method to identify the relevance of diseases in the whole-genome. Although whole-genome microarray technology can detect more than 4 million markers per sample, it is only effective for known mutations, and detection of new mutations is not ideal. Next-generation sequencing (NGS) technology provides a more comprehensive method, which is an unbiased mutation detection method. NGS does not consider the prior expectations, and detects the 3.2 billion pairs of bases in the human genome one by one. For example, whole-exome and whole-genome sequencing are commonly used methods for detecting pathogenic variants in rare or complex disease research.
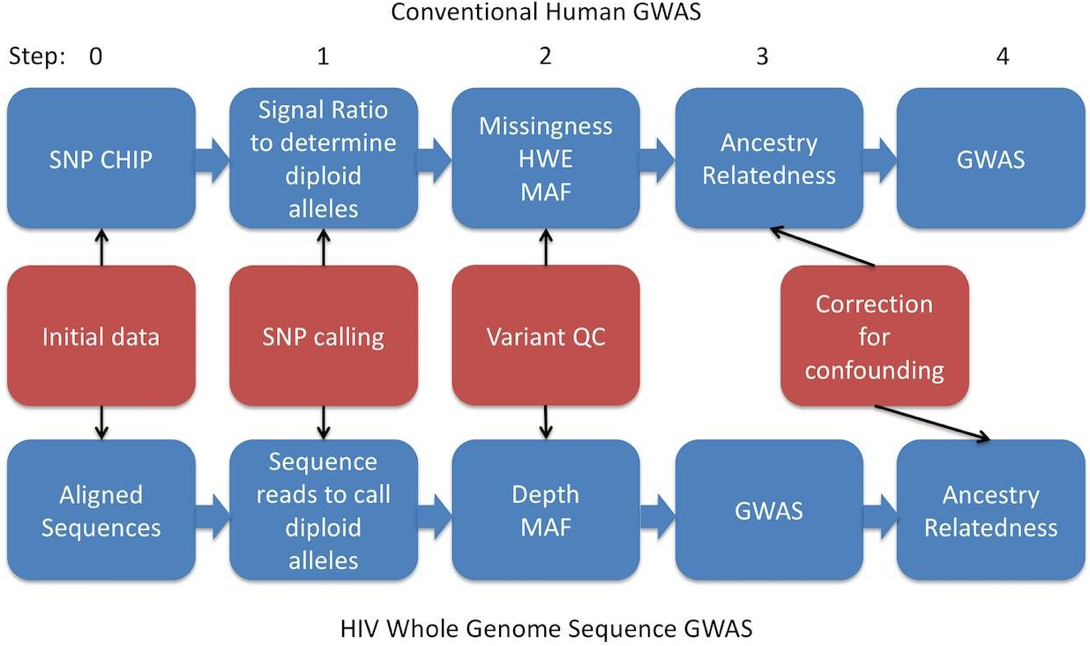 Fig 1. Analysis pipeline for HIV whole-genome sequence (WGS) genome-wide association study (GWAS) compared to human research using an SNP chip. (Power R A,et al. 2016)
Fig 1. Analysis pipeline for HIV whole-genome sequence (WGS) genome-wide association study (GWAS) compared to human research using an SNP chip. (Power R A,et al. 2016)
Advantages of Whole-Genome SNP Genotyping
The whole-genome genotyping method is widely used in basic disease research. Compared with microarray chips, the whole-genome genotyping method has the following advantages:
- Provide a comprehensive view of the genome.
- Detect single nucleotide polymorphisms (SNP) and other variants in the whole-genome.
- Potential pathogenic variants can be identified for future targeted research.
- Minimize content restrictions and expand the scope of exploration.
Applications
The main applications of whole-genome SNP genotyping analysis in biomedical are:
- Population research: Genome-wide SNP typing helps laboratories to carry out population-scale genotyping research to detect common genetic variations in large-scale populations.
- Basic disease research: Quickly screen a large number of samples to find pathogenic variants related to complex diseases.
What We Offer
As one of the global biological information analysis service providers, CD Genomics provides established, cost-efficient, and rapid turnaround analysis services for whole-genome SNP genotyping analysis for researchers, aiming to help you understand the genotype of each gene locus from the whole-genome level. CD Genomics provides personalized whole-genome SNP genotyping analysis services to meet the analysis needs of different customers. In addition, we are able to receive various formats of data (such as whole-genome data, whole exome data and microarray data) for analysis such as raw data files, or other intermediate data formats. We will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis result report.
Data Analysis Technical Route
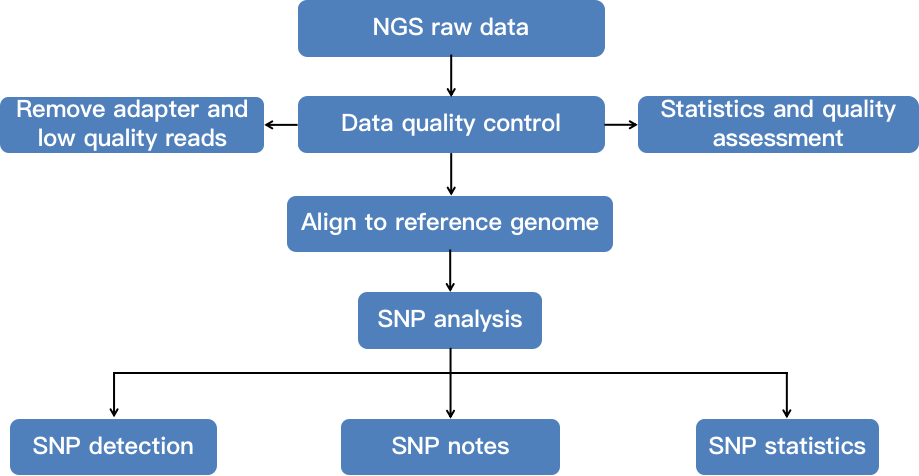 Fig 2. A flow chart demonstrating the whole-genome SNP genotyping analysis
Fig 2. A flow chart demonstrating the whole-genome SNP genotyping analysis
Data Ready
Before data analysis, the first thing is to get your data ready. For whole-genome SNP genotyping analysis, the input data can be microarray data or NGS data, which can be obtained from the following channels:

To process data more efficiently, we prefer to receive data files in the raw format, but we can also receive pre-normalized files. More importantly, there are currently many databases related to disease whole-genome data or whole exome data, we are able to provide services for obtaining and mining data from available databases. Alternatively, if you do not currently have the input data, CD Genomics can also provide you with a variety of microarray or sequencing services based on its rich experience. If you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
Biomedical-Bioinformatics, as a division of CD Genomics, provides a one-stop whole-genome SNP genotyping analysis service for biomedical researchers. With years of experience in biological data analysis, CD Genomics' analysis engineers will provide you with the most appropriate analysis strategy based on your data, and generate high-quality results and charts that can be used for publication. In addition to whole-genome SNP genotyping analysis, we also provide other genotyping analysis based on SNP chip services. For more detailed information, such as microarray or sequencing services and other data analysis services, please feel free to contact us.
References
- Power R A, et al. Genome-Wide Association Study of HIV Whole-Genome Sequences Validated using Drug Resistance[J]. Plos One, 2016, 11(9).
- Myoungsook, et al. Genome-wide association study for the interaction between BMR and BMI in obese Korean women including overweight [J]. Nutrition Research & Practice, 2016. Feb; 10(1): 115–124.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
