Residue Interaction Network
We provide customers with high-quality residue interaction network analysis that aims to help researchers or doctors study molecular mechanisms of diseases and drug design from a systematic perspective.
Introduction
Proteins are composed of 20 different amino acids polymerized by peptide bonds. They form specific tertiary structures to catalyze and regulate different biological functions. Different amino acid arrangements will form different protein structures. The three-dimensional structure of the protein determines its biological function. Therefore, the study of protein three-dimensional structure is of great significance for understanding how it exerts its biological functions and designing related drugs. Among them, protein residue interaction network analysis is widely used in the study of protein-related problems. In this method, the nodes of the network are the residues that make up the protein, and the edges of the network are the interactions of non-covalent residues, such as van der Waals and electrostatic interactions. Based on the interaction network of protein residues, graph theory can be further used to study protein structure stability, protein dynamics, enzyme activity and allosteric regulation, signal transduction and other issues. The interaction network of protein residues plays an important role in understanding the structural characteristics of proteins, biological functions, and prediction of binding sites. It is of great help to disease research and drug design-related application research in the field of biomedicine.
An Example of Residue Interaction Network Analysis Result
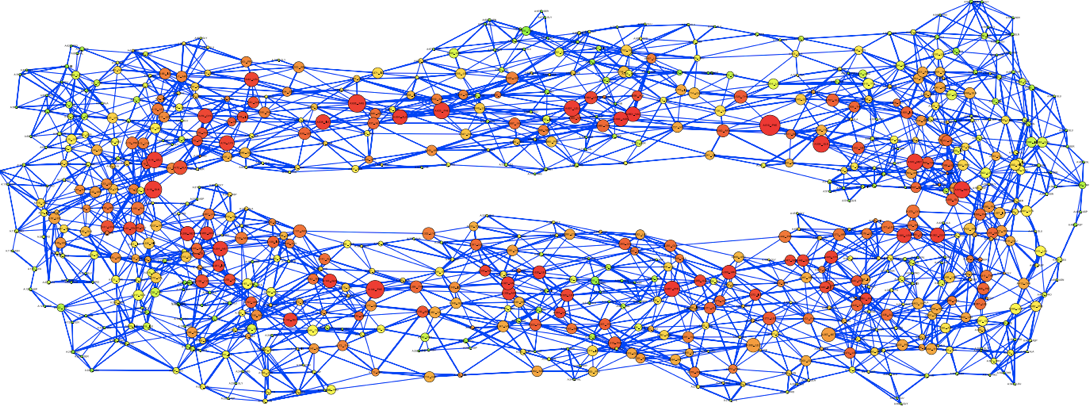 Fig 1. The residue interaction network for the β clamp in the research of residue interaction network analysis of Dronpa and a DNA clamp. (Hu G, et al. 2014)
Fig 1. The residue interaction network for the β clamp in the research of residue interaction network analysis of Dronpa and a DNA clamp. (Hu G, et al. 2014)
Application Fields
In the field of biomeBiomedical-Bioinformatics, a division of CD Genomics, provides customers with high-quality residue interaction network analysis, which aimsdicine, the residue interaction network analysis is mainly used in disease and drug design research:
- Protein structure research: Protein structure stability, protein dynamics, enzyme activity and allosteric regulation, signal transduction.
- Protein biological function research: Protein biological function and binding site prediction, disease occurrence and development mechanism research, and drug design analysis.
What We Offer
As one of the global bioinformatics data analysis service providers, CD Genomics provides established, cost-efficient, and rapid turnaround analysis services for residue interaction network analysis for doctors and researchers. CD Genomics provides different software or tools such as AMINONET, RING, Cytoscape, NAPS, and other cutting-edge software to perform residue interaction network analysis and visual analysis for researchers to meet the personalized needs. In addition, you only need to provide us with your original data or data analysis requirements. We will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis We will generate high-quality charts that can be directly used for publication. We provide researchers with the following data analysis services:
- Communicate the residue interaction network analysis needs.
- Develop an analysis plan (including the selection of analysis strategies and software).
- Residue interaction network analysis and personalized analysis content.
- Generate complete results and reports.
- Repeatable analysis results and fast analysis process.
Data Analysis Technical Route
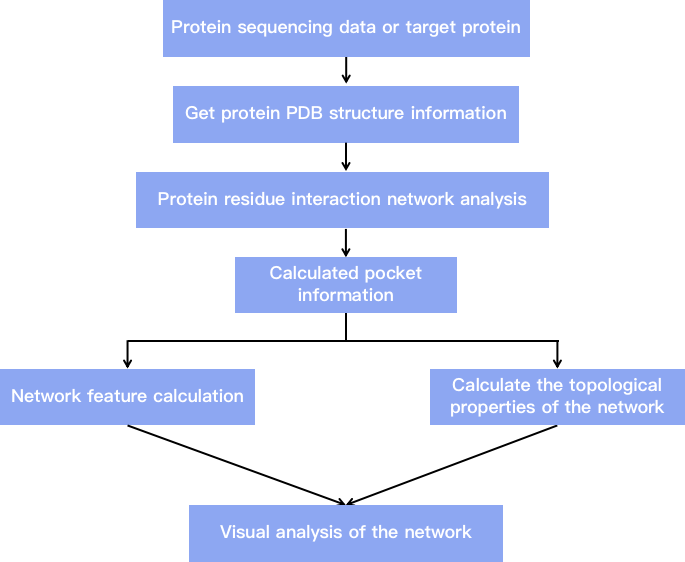 Fig 2. Flow chart showing our residue interaction network analysis
Fig 2. Flow chart showing our residue interaction network analysis
Service Process
Before data analysis, the first thing is to get your data ready. For residue interaction network analysis, the raw input data can be protein sequencing raw data or target protein file. For protein sequencing raw data, we first identify and analyze the protein, and then provide appropriate data analysis services based on your data. Our service process is as follows:
-
Upload protein sequencing raw data or target protein file
-
Develop an analysis plan
-
Residue interaction network analysis
-
Generate charts
-
After-sales service
For residue interaction network analysis services, if you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
With years of experience in biological data analysis, CD Genomics analysis engineers will provide you with the most appropriate analysis strategy based on your data, and generate high-quality results and charts that can be used for publication. For more detailed information about our data analysis services, please feel free to contact us.
References
- Hu G, et al. Residue interaction network analysis of Dronpa and a DNA clamp[J]. Journal of Theoretical Biology, 2014, 348:55-64.
- Griffin J, et al. Residue Interaction Network Analysis Predicts a Val24–Ile31 Interaction May be Involved in Preventing Amyloid〣eta (1–42) Primary Nucleation[J]. The Protein Journal, 2021:1-9.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
