Molecular Diagnostic Analysis of Infectious Diseases
With years of experience in metagenomic data analysis, CD Genomics provides different types of metagenomics next-generation sequencing (mNGS) data analysis (such as targeted mNGS data analysis and untargeted mNGS data analysis) services that can help researchers identify and analyze pathogens in infectious disease research. In addition, it also identifies unknown pathogens or known pathogens with large variations, and at the same time brings new ideas for molecular diagnosis of infectious diseases and the prevention and control of infectious diseases.
Introduction
Metagenomics research is widely used in soil, water and other environmental samples, as well as the biodiversity analysis of microbial communities related to human diseases. Since metagenomics does not require separate isolation and culture of pathogens (such as viruses, bacteria, fungi and/or parasites), clinical samples can be directly analyzed through nucleic acid extraction and purification, which provides new technical means and ideas for the detection of infectious disease pathogens, especially unknown pathogens. Metagenomics-based second-generation sequencing (mNGS) can obtain the genomes of all species in the sample. Through bioinformatics analysis technology, it can identify all ppathogens at the same time based on uniquely identifiable DNA and/or RNA sequences. It greatly reduces the time, manpower and material resources for eliminating suspicious pathogens one by one, and can identify unknown pathogens or pathogens with known mutations, bringing new ideas for the prevention and control of infectious diseases. In addition, another key advantage of the NGS method is that the sequencing data may be used for other analysis besides identifying pathogenic agents, such as RNA sequencing (RNA-seq) to analyze the transcriptome to determine the characteristics of the microbiome and humans. At present, mNGS has been widely used in public health surveillance, pathogen detection, and traceability of infectious diseases.
Metagenomics Bioinformatics Analysis Pipeline in Infectious Disease Research
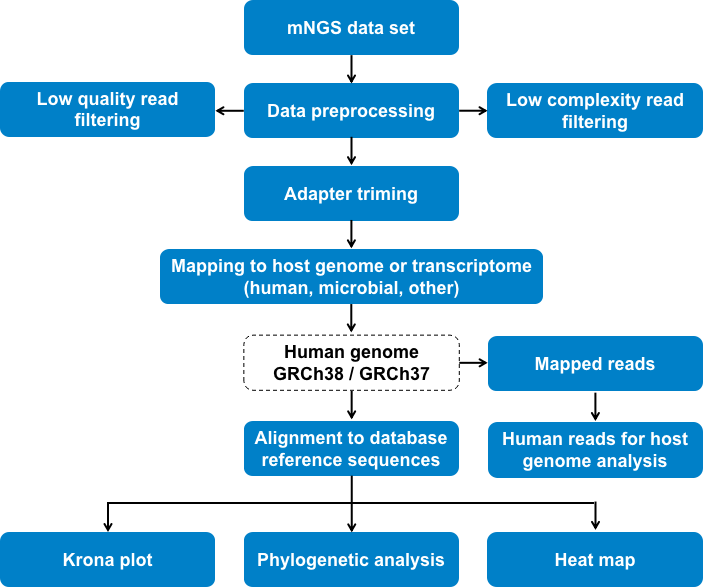 Fig 1. Metagenomics next-generation sequencing (mNGS) bioinformatics analysis pipeline in infectious disease research.
Fig 1. Metagenomics next-generation sequencing (mNGS) bioinformatics analysis pipeline in infectious disease research.
Application of mNGS data analysis in the Diagnosis of Infectious Diseases
- Pathogen monitoring: Metagenomic data analysis can overcome the limitations of traditional methods, achieve high-precision identification of pathogens, and provide guidance for daily pathogen monitoring.
- Pathogen detection: Sequencing data analysis based on metagenomics can detect unknown pathogens and assist clinical diagnosis.
- Pathogen tracing: When investigating the epidemic situation of infectious diseases, metagenomics can trace the pathogen after sequencing and analyzing samples, find the potential transmission route, and identify and cut off the source of infection in time.
- Efficient control of emerging infectious diseases: Rapid analysis of infectious disease pathogens to find the source and control emerging infectious diseases.
What We Offer
CD Genomics relies on years of experience in sequencing and bioinformatics data analysis, provides doctors and researchers with established, cost-efficient, and rapid turnaround data analysis services of mNGS data analysis in the research of infection disease.
Targeted Metagenomics NGS Analysis: In the molecular diagnostic analysis of infectious diseases, targeted detection methods for next-generation metagenomics sequencing are beneficial to increase the number and proportion of pathogen reads in sequence data, and can improve the detection sensitivity of target microorganisms.
LEARN MOREUntargeted Metagenomics NGS Analysis: Untargeted metagenomic sequencing requires unbiased shotgun sequencing of all microorganisms and host nucleic acids in clinical samples. It can be used to analyze various patient samples and cultured microbial colonies for pathogen identification, microbiome analysis, and/or host transcriptome analysis.
LEARN MOREWhat's More
Biomedical-Bioinformatics, a division of CD Genomics, provides customers with one-stop mNGS data analysis services in the molecular diagnostic analysis of infectious diseases. In addition to data analysis services, we can also provide cost-effective sequencing services and services for downloading raw data from the database according to the analysis needs of customers. For data analysis services, if you have any questions, please feel free to contact our professional technical support. We are always ready to provide you with satisfactory services.
References
- Gu W, et al. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection[J]. Annu Rev Pathol. 2019 Jan 24; 14:319-338.
- Chiu C Y, Miller S A. Clinical metagenomics[J]. Nature Reviews Genetics, 2019. Jun;20(6):341-355.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.