SV Analysis
Introduction
Structural variants (SVs) are genomic rearrangements affecting more than 50 bp. Genomic structure variation (SV for short) usually refers to large-length sequence changes and positional changes in the genome. There are many types of genomic structure variation, including insertion or deletion of long fragment sequences longer than 50bp (Big Indel), Tandem repeat, chromosome inversion (Inversion), sequence translocation within or between chromosomes (Translocation), copy number variation (CNV) and more complex chimeric variation. Commonly used methods for structural variation detection include:
- Array-based detection: Including microarray comparative genome hybridization (array CGH).
- Detection methods based on high-throughput sequencing: Such as whole genome resequencing (WGS), simplified genome sequencing, CLR detection, HiFi mutation detection, etc.
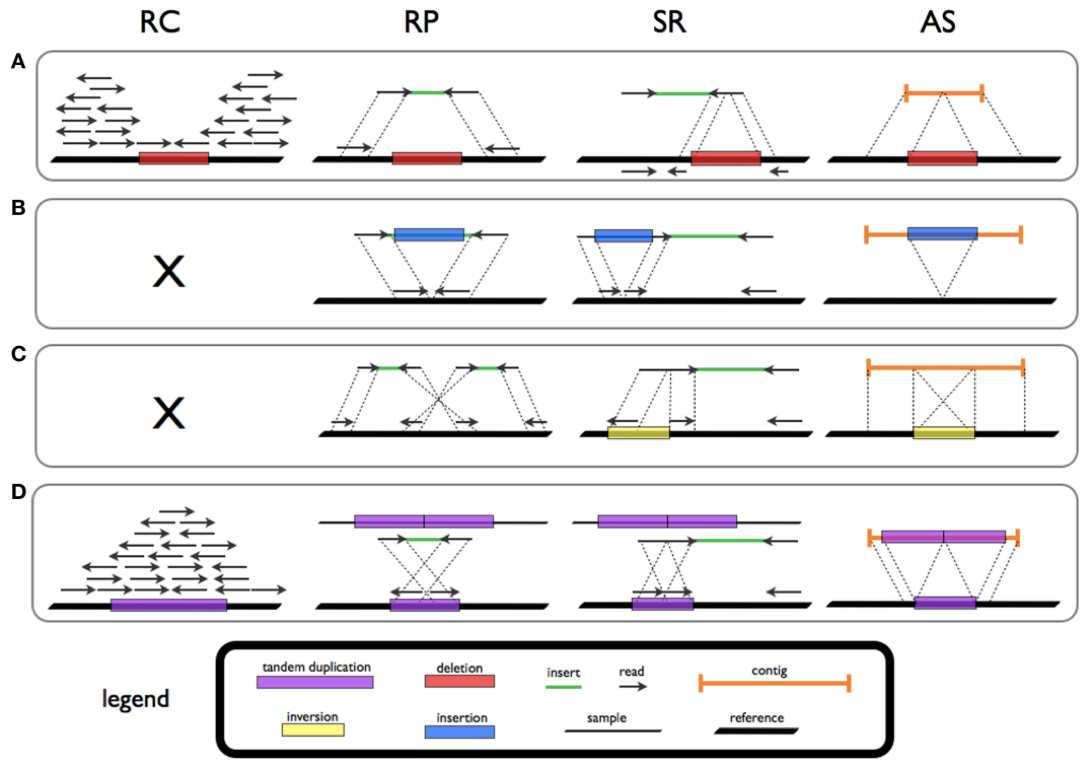 Fig 1. Patterns and signatures of SVs for deletion (A), novel sequence insertion (B), inversion (C), and tandem duplication (D) in read count (RC), read-pair (RP), split-read (SR), and de novo assembly (AS) methods. (Tattini L, et al. 2015)
Fig 1. Patterns and signatures of SVs for deletion (A), novel sequence insertion (B), inversion (C), and tandem duplication (D) in read count (RC), read-pair (RP), split-read (SR), and de novo assembly (AS) methods. (Tattini L, et al. 2015)
The Importance of SV Analysis
SV analysis is often used in the research of various diseases:
1. SVs have a greater impact on the genome than SNPs, and once they occur, they will often have a significant impact on the living body, such as causing birth defects, cancer, etc.
2. Studies have found that SVs on the genome are more representative of the diversity of the human population than SNPs.
3. Some structural variants that are rare and the same are often related to the occurrence of diseases (including cancer) and even are their direct causes.
What We Offer
As one of the experienced biomedical data analysis service providers, CD Genomics offers established, cost-efficient, and rapid turnaround analysis services for SV analysis for researchers. CD Genomics provides different software such as Manta, Delly, AS-GENSENG, BICseq2, CNVnator, BreakDancer, Breakway, CLEVER to perform SV analysis for customers to meet the analysis needs of different research directions. In addition, CD Genomics provides customers with personalized analysis services. The raw input data can be microarray data or different types of high-throughput sequencing data. In addition, we are able to receive various formats of data for analysis such as raw data files, or other intermediate data formats. We provide our clients with the following services:
- Data analysts evaluate and filter raw data.
- Develop suitable analysis strategies according to the data situation.
- Perform structural variants analysis.
- Provide a complete report summarizing the results and report interpretation services.
- Experienced bioinformatics engineers complete all analysis content in a short period.
Data Analysis Technical Route
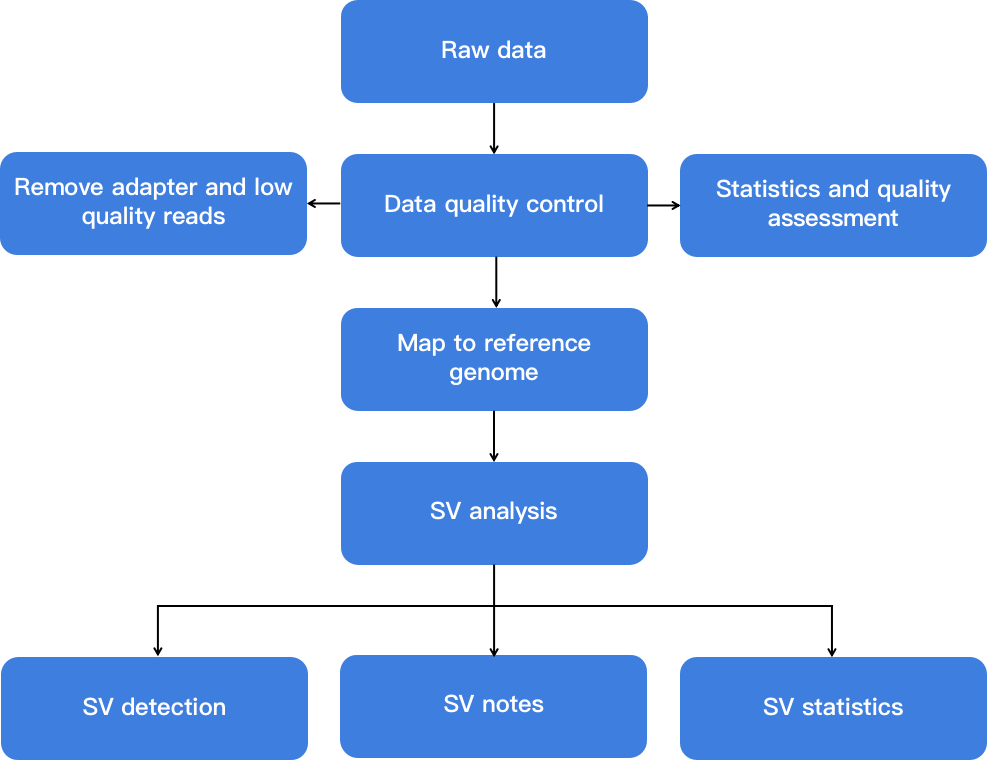 Fig 2. Flow chart showing SV analysis.
Fig 2. Flow chart showing SV analysis.
Data Ready
Before data analysis, the first thing is to get your data ready. Data can be obtained from the following channels:
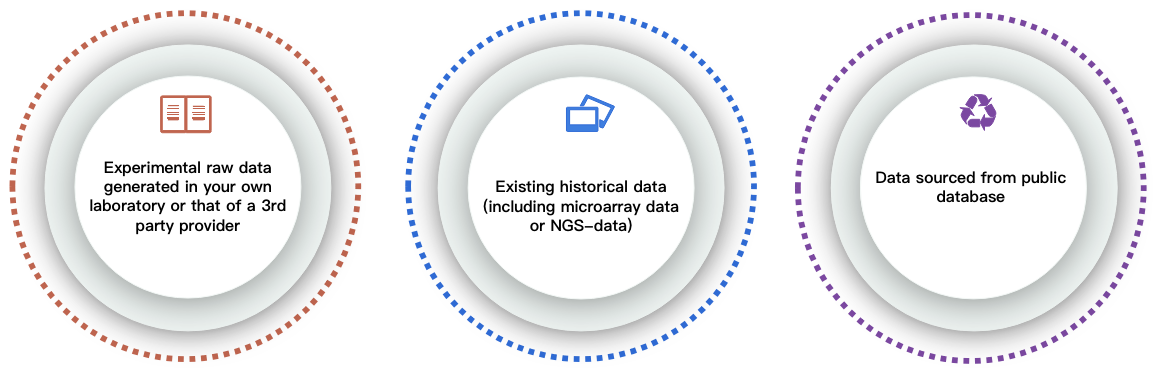
In order to process data more efficiently, we prefer to receive data files in the raw format, but we can also accept pre-normalized files. More importantly, if you do not currently have the original data, CD Genomics can also provide you with a variety of sequencing services based on its rich sequencing experience. Alternatively, there are currently many databases related to SV, we are able to provide services for obtaining and mining data from available databases. If you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
Biomedical-Bioinformatics, as a division of CD Genomics, provides customers with one-stop gene structure variation analysis services based on its rich data analysis experience. In addition, it also provides other types of variation analysis services such as CNV analysis, InDel analysis, SNP fine mapping, etc. For more detailed information, such as sequencing services and SV data analysis services, please feel free to contact us.
References
- Tattini L, et al. Detection of Genomic Structural Variants from Next-Generation Sequencing Data[J]. Frontiers in Bioengineering and Biotechnology, 2015: 92-92.
- Kosugi S, et al. Comprehensive evaluation of structural variation detection algorithms for whole genome sequencing[J]. Genome Biology, 2019, 20(1).
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
