Clinical Metagenomics Analysis
With the advancement of high-throughput sequencing library preparation methods, sequence generation and computational bioinformatics analysis technology, it is possible to conduct faster and more comprehensive metagenomic analysis at a lower cost. As a new type of microbial detection method, metagenomics next-generation sequencing (mNGS) may replace the current culture, antigen detection and PCR methods in clinical microbiology in the future. With years of experience in microbial data analysis, CD Genomics provides customers with comprehensive and high-quality metagenomics data analysis services to accelerate your metagenomics-related research.
Introduction
The metagenome is the sum of the genetic material of all the tiny organisms in the habitat. It contains the genomes of cultivable and non-cultivable microorganisms. Metagenomics takes the microbial population genome in environmental samples as the research object, and uses sequencing technology and bioinformatics analysis technology as research methods. Research on microbial diversity, population structure, evolutionary relationship, functional activity, mutual cooperation and the relationship between microorganisms and the environment in order to interpret the diversity and abundance of microbial populations, explore the relationship between microbes and the environment, and between microbes and hosts, discover and study new genes with specific functions. Clinical metagenomics next-generation sequencing (mNGS) is an emerging diagnostic technology for the comprehensive analysis of microorganisms and host genetic material (DNA and RNA) in patient samples. This emerging method is changing the way doctors diagnose and treat diseases. It has a wide range of applications, including antimicrobial resistance research, microbiome, human host gene expression (transcriptomics), and oncology.
Metagenomics Data Analysis Process
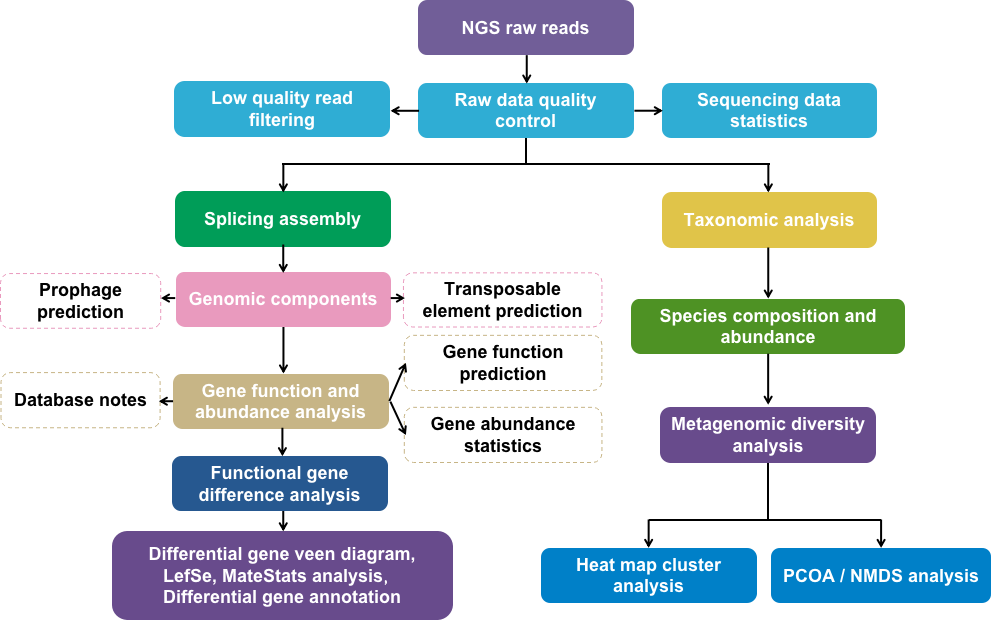 Fig 1. Metagenomics next-generation sequencing (mNGS) bioinformatics data analysis process.
Fig 1. Metagenomics next-generation sequencing (mNGS) bioinformatics data analysis process.
Application of Clinical Metagenomics Research
1. Pathogen identification: Such as pathogen identification on samples of various infectious diseases and syndromes.
2. Pathogenic microbiome research: Analysis of the microbiome in disease and health conditions, transcriptomics analysis of human hosts after infection with pathogens, and identification of tumor-associated viruses and their genome integration sites.
3. Public health: Identify pathogens from clinical samples to guide the management and treatment strategies of infected patients, public health microbiology, and monitor the source of infectious disease outbreaks in the community.
What We Offer
CD Genomics relies on years of experience in metagenomics sequencing and bioinformatics data analysis to provide doctors and researchers with established, cost-efficient, and rapid turnaround data analysis services of mNGS data analysis in different research areas, such as infectious disease diagnosis, clinical microbiology, human host gene expression (such as transcriptomics), oncology and antimicrobial resistance research and other fields.
Molecular Diagnostic Analysis of Infectious Diseases: In the molecular diagnostic analysis of infectious diseases, targeted metagenomics NGS analysis or untargeted metagenomics NGS analysis method is used to analyze various patient samples and cultured microbial colonies for pathogen identification and microbial composition analysis.
- Targeted Metagenomics NGS Analysis
- Untargeted Metagenomics NGS Analysis
What's More
Biomedical-Bioinformatics, a division of CD Genomics, provides customers with one-stop clinical metagenomics next-generation sequencing data analysis services in different fields of pathogen research. In addition, for clinical metagenomics research, if you have yet to generate the raw sequencing data, CD Genomics can help by arranging for the generation of your data. We also have a variety of sequencing platforms, including PacBio, Nanopore, Illumina, and Miseq, etc. If you have any questions about our services, please feel free to contact our professional technical support. We are always ready to provide you with satisfactory services.
References
- Chiu C Y, Miller S A. Clinical metagenomics[J]. Nature Reviews Genetics, 2019. Jun;20(6):341-355.
- Zhao F, Bajic VB. The value and significance of metagenomics of marine environments[J]. Genomics Proteomics Bioinformatics. 2015; 13:271–274.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.