LncRNA and Metabolome Integration Analysis
Introduction
Long non-coding RNA (lncRNA) is a class of molecules that are longer than 200 nucleotides but do not encode proteins. LncRNA is ubiquitous in humans and various eukaryotes. It has the characteristics of extensive transcription, many types, and diverse functions. It is an essential regulator of various biological processes and plays a regulatory role through various mechanisms. Metabolome is the final result of gene transcription under internal and external regulation of organisms, and is the direct manifestation of the material basis and phenotype of organisms.
In the era of systems biology research, biological development processes are complex and changeable, and gene regulatory networks are complex. Researching systems biology with a single omics is often one-sided. The correlation analysis between lncRNA sequencing data and metabolome data can go deep into regulatory and functional genes, analyze the internal changes of organisms from two levels of cause and effect, lock key pathways with metabolite changes, and build a core regulatory network, and then systematically and comprehensively analyze the complex mechanism of disease occurrence and development, and explain biological problems as a whole.
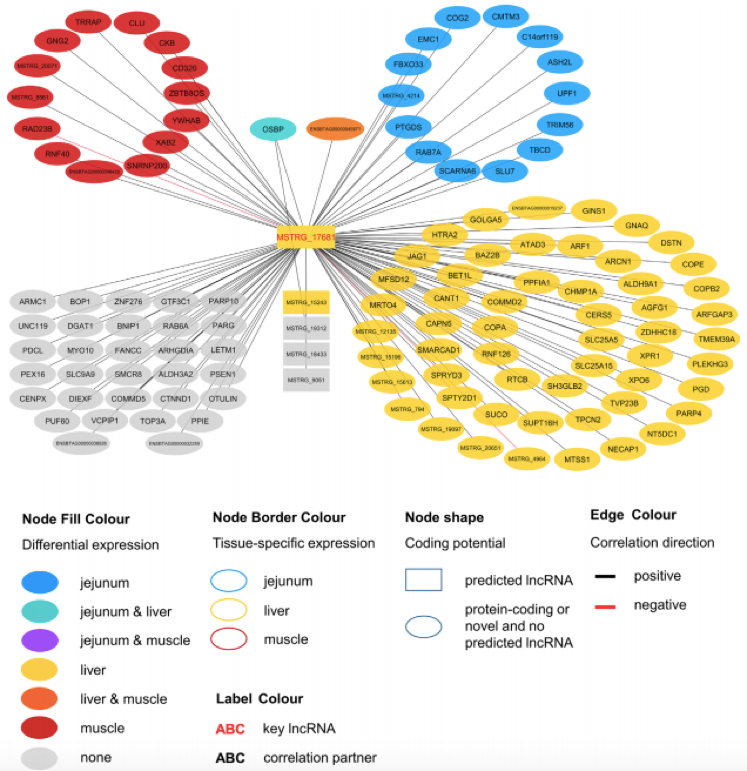 Figure1. Co-expression network for the novel long non-coding RNA MSTRG.17681 with key regulatory potential for metabolic efficiency in cattle and significantly (p < 0.05) correlated genes with a minimal correlation coefficient of |r| > 0.8. (Nolte W, et al. 2019)
Figure1. Co-expression network for the novel long non-coding RNA MSTRG.17681 with key regulatory potential for metabolic efficiency in cattle and significantly (p < 0.05) correlated genes with a minimal correlation coefficient of |r| > 0.8. (Nolte W, et al. 2019)
Application Fields
LncRNA and metabolome integration analysis is used in many fields of biomedical research:
- Basic disease research: Research on the regulation mechanism of disease occurrence and development and biomarker discovery.
- Clinical diagnosis: disease classification, personalized treatment.
What We Offer
CD Genomics provides a one-stop bioinformatics data analysis service for doctors and medical researchers, and provides researchers with mature, cost-effective and fast turnaround analysis services for lncRNA and metabolome integrated analysis. The raw input lncRNA data and metabolome data can be produced from different sequencing platforms or mass spectrometers and we can use various formats of data for analysis such as raw data files, or other intermediate data formats. We provide researchers with the following services:
- Raw data filtering and evaluation.
- Develop an analysis plan (including the selection of analysis strategies and software).
- Long non-coding RNA and metabolome integration analysis.
- Generate a comprehensive report.
- Repeatable analysis results and fast analysis process.
Data Analysis Technical Route
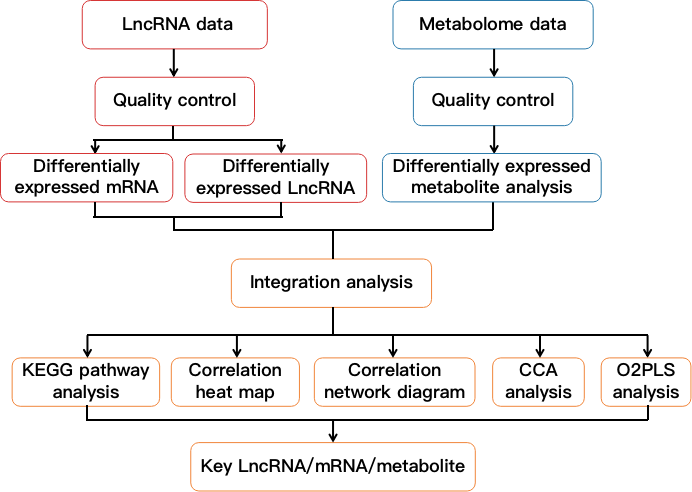 Fig 2. Flow chart showing lncRNA and metabolome integration analysis.
Fig 2. Flow chart showing lncRNA and metabolome integration analysis.
Analysis Content Includes:
Based on lncRNA sequencing data and metabolome data, we first perform differentially expressed analysis (including lncRNA, mRNA and metabolite), and then perform integration analysis on the differentially expressed substances. The content of the correlation analysis is as follows:
- Enrichment analysis of KEGG pathway.
- Correlation heat map.
- Correlation network diagram.
- Canonical correspondence analysis (CCA analysis).
- O2PLS model correlation analysis.
- Identify key LncRNA/mRNA/metabolites.
Data Ready
Before data analysis, the first thing is to get your data ready. The raw data or intermediate data can be obtained from the following channels:
In order to process data more efficiently, we prefer to receive data files in the raw format, but we can also accept pre-normalized files. In addition, if you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
For lncRNA and metabolome integration analysis, if you don't have the raw data, CD Genomics provides you with sequencing services, and we will formulate a suitable lncRNA and metabolome sequencing program according to your research purposes. Alternatively, we are able to provide services for obtaining and mining data from different databases. For more detailed information, please feel free to contact us.
Reference
- Nolte W, et al. Biological Network Approach for the Identification of Regulatory Long Non-Coding RNAs Associated with Metabolic Efficiency in Cattle[J]. Frontiers in Genetics, 2019, 10: 01130.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
