InDel Analysis
Introduction
InDel refers to the insertion or deletion sequence of small fragments in the genome, and its length is between 1-50 bp. The reason is that the read length of Illumina sequencing is about 100 bp, including single-end sequencing (100 bp) and paired-end sequencing (2 x 100 bp). Therefore, in SNP calling, most of the reliable indels that can be detected are less than 100 bp, and usually the maximum is about 50 bp. InDel variation is generally less than SNP variation, which also reflects the difference between the sample and the reference genome. InDel in the coding region will cause frameshift mutations, leading to changes in gene function. Commonly used methods for structural variation detection include:
- High-throughput sequencing-based detection methods: Such as simplified genome sequencing, whole genome resequencing (WGS), CLR detection, CCS HiFi mutation detection, etc.
- Array-based detection: Including microarray comparative genome hybridization (array CGH).
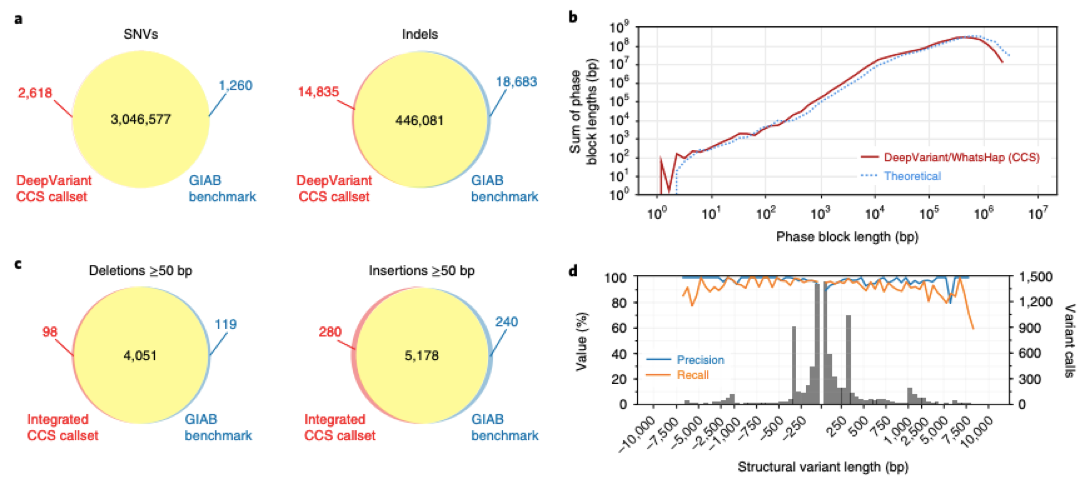 Fig 1. Variant calling (including SNVs, indels, deletions, and insertions) and phasing with CCS reads. (Wenger A M, et al. 2019)
Fig 1. Variant calling (including SNVs, indels, deletions, and insertions) and phasing with CCS reads. (Wenger A M, et al. 2019)
What We Offer
As one of the global bioinformatics analysis service providers, CD Genomics provides established, cost-efficient, and rapid turnaround analysis services for indel analysis for researchers, aiming to help you detect the indels in different samples. CD Genomics provides different software such as GATK, samtools, TASSEL, DeepVariant to perform indel analysis for customers to meet the analysis needs of different research directions. In addition, we can use various formats of data for analysis such as raw data files, or other intermediate data formats. You only need to provide us with your original data, and we will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis result report. We provide our clients with the following services:
- Professional data analysts evaluate and filter the data, formulate the optimal analysis plan, and perform data analysis.
- Develop a personalized analysis plan, or develop a personalized result chart according to the analysis needs of customers.
- Provide a complete interactive data analysis report, including all analysis methods and results.
- Post-report follow-ups: We provide a professional analysis report interpretation service and biological interpretation of analysis results.
- Fast turnaround time.
Data Analysis Technical Route
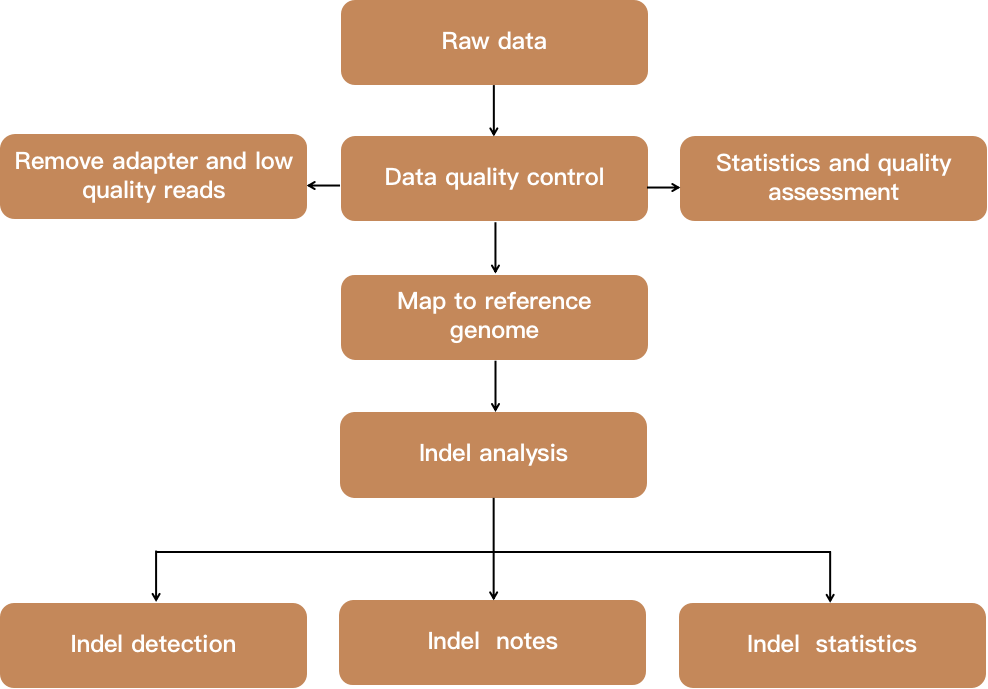 Fig 2. Flow chart showing indel analysis.
Fig 2. Flow chart showing indel analysis.
An Example of Indel Analysis Process
Different types of raw data will have different analysis schemes. The following is the general process of indel analysis using RNA-seq data (use GATK to detect indels):
1. Use STAR software to compare the data to the reference genome (mapping to the reference).
2. Use Picard's "MarkDuplicates" command to perform data cleanup.
3. GATK's SplitNCigarReads package is used to process reads containing N in cigar.
4. Base Quality Recalibration (Base Quality Recalibration) is to use machine learning to adjust the quality score of the original base.
5. Variant calling, filtering and annotation.
Data Ready
Before data analysis, the first thing is to get your data ready. For indel analysis, the raw input data can be microarray data or different types of high-throughput sequencing data, and the data can be obtained from the following channels:
To process data more efficiently, we prefer to receive data files in the raw format, but we can also accept pre-normalized files. More importantly, there are currently many databases related to indel. We are able to provide services for obtaining and mining data from available databases. Alternatively, if you do not currently have the input data, CD Genomics can also provide you with a variety of sequencing services based on its rich sequencing experience. If you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
Biomedical-Bioinformatics, as a division of CD Genomics, provides customers with one-stop gene structure variation analysis services based on its rich data analysis experience. In addition to indel analysis, it also provides other types of variation analysis services such as CNV analysis, SV analysis, SNP fine mapping, etc. For more detailed information, such as sequencing services and indel data analysis services, please feel free to contact us.
Reference
- Wenger A M, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome[J]. Nature Biotechnology, 2019, 37(11).
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
