Untargeted Metagenomics NGS Analysis
With years of experience in microbial data analysis, CD Genomics provides you with comprehensive targeted meta-genomics next-generation sequencing (mNGS) data analysis and targeted mNGS data analysis services, and through the analysis of different primitive types of data, in-depth mining of pathogen information related to infectious diseases.
Introduction
Compared with targeted mNGS, which specifically increases the number and proportion of sequence data of target microorganisms, untargeted mNGS analysis does not use specific primers or probes to amplify samples during the library building process. Thus, the DNA and/or RNA in the entire sample can be sequenced. Untargeted metagenomic sequencing requires unbiased shotgun sequencing of all microorganisms and host nucleic acids in clinical samples. Separate DNA and RNA (reversed to cDNA) libraries are constructed. The DNA library is used to identify bacteria, fungi, DNA viruses and parasites, and the RNA library is used to identify RNA viruses and human host transcriptome analysis based on RNA sequencing. Untargeted next-generation metagenomic sequencing (mNGS) methods can be used to analyze various patient samples and cultured microbial colonies for pathogen identification, microbiome analysis, and/or host transcriptome analysis. Almost all pathogens, including viruses, bacteria, fungi and parasites, can be identified in a single test.
Untargeted Metagenomics Next-Generation Sequencing Pipeline
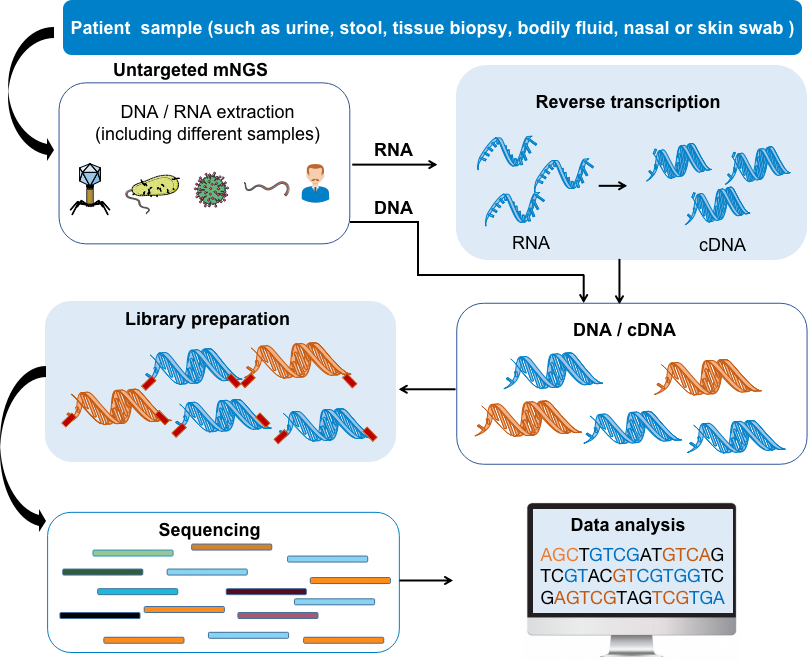 Fig 1. Untargeted meta-genomics next-generation sequencing (mNGS) process.
Fig 1. Untargeted meta-genomics next-generation sequencing (mNGS) process.
Untargeted Metagenomic NGS Analysis Application Areas
- Identify the types of pathogenic microorganisms and assist in the diagnosis of clinical diseases.
- Clinical pathogen genome assembly.
- Pathogen monitoring, such as pathogen subtype analysis and variation tracking.
- With the analysis of the diversity of pathogenic microorganisms in different samples, the clinical sample data is enriched to characterize the analysis of antibiotic resistance.
What We Offer
CD Genomics relies on years of experience in sequencing and bioinformatics data analysis, provides doctors and researchers with established, cost-efficient, and rapid turnaround data analysis services of untargeted metagenomics next-generation sequencing data analysis in the research of infection disease. CD Genomics provides different software or packages to perform mNGS data analysis and visual analysis for researchers to meet their personalized needs. In addition, you only need to provide us with your original NGS data or analysis requirements, we will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis report, with high-quality results and charts that can be directly used for article publication.
Data Ready
Before data analysis, the first task is to upload raw data. We will evaluate the quality of the original data, and then formulate an appropriate analysis plan. The original data can be from any of the following channels:
For untargeted meta-genomics data analysis services, if you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
CD Genomics provides customers with one-stop data analysis services, and what customers need to do is to provide raw data and analysis requirements, CD Genomics will develop corresponding analysis strategies based on customer data and provide high-quality reports. Importantly, for untargeted meta-genomics next-generation sequencing data analysis, except data analysis services, we can also provide customers with corresponding targeted genome high-throughput sequencing services and downloading data from public databases. In addition to untargeted metagenomics NGS analysis, we also provide targeted metagenomics NGS analysis services. If you have any questions about our services, please feel free to contact us for details.
References
- Chiu C Y, Miller S A. Clinical metagenomics[J]. Nature Reviews Genetics, 2019. Jun;20(6):341-355.
- Gu W, et al. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection[J]. Annu Rev Pathol. 2019 Jan 24; 14:319-338.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
