Non-coding RNA Regulation Analysis
With years of experience in non-coding RNA data analysis services, CD Genomics provides scientists with fast, efficient and accurate one-stop non-coding RNA regulation analysis services.
Introduction
Non-coding RNA (ncRNA) is an RNA molecule that does not code for the protein transcribed from the genome. In addition to playing a role at the transcriptional and post-transcriptional level, non-coding RNA also plays an important role in the epigenetic regulation of gene expression. In the genome of eukaryotes, approximately 90% of the genes are transcribed genes. Only 1-2% of these transcribed genes encode proteins, and most of the others are transcribed as non-coding RNA. Non-coding RNA can be divided into two main types: basic structural ncRNA and regulatory ncRNA. Basic structural ncRNAs play a role similar to housekeeping genes in translation and splicing, and include species such as ribosomal RNA (rRNA), transfer RNA (tRNA), and small nuclear RNA (snRNA), etc. Regulatory ncRNAs mainly include the following categories: microRNA (miRNA), Piwi-interacting RNA (piRNA), small interfering RNA (siRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA), etc.
Classification of Non-coding RNAs
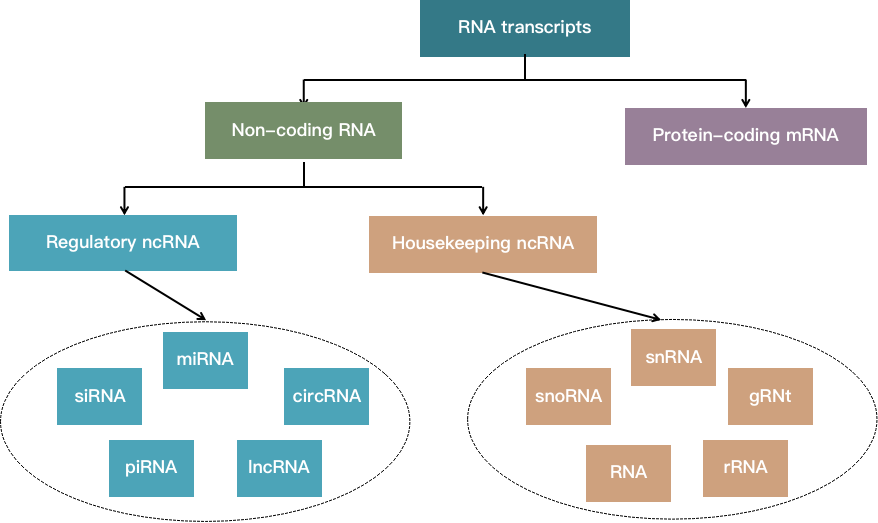 Fig 1. Classification of non-coding RNAs based on their functions.
Fig 1. Classification of non-coding RNAs based on their functions.
Functions of Regulatory Non-coding RNAs
- microRNA (miRNA): miRNA is a small single-stranded molecule with a length of about 20-24 nt. It is derived from a transcript that forms a unique hairpin structure. It is called pre-miRNA and participates in post-transcriptional gene expression regulation. The miRNA will pair with the complementary sequence on the target mRNA transcript through the 3'UTR, resulting in the silencing of the target gene.
- Piwi interaction RNA (piRNA): piRNA is a small molecule ncRNA (24-31nt) that can form a complex with the Piwi protein of the Argonaute family. piRNA is characterized by uridine at the 5'end and 2'-O-methyl modification at the 3'end. It is mainly combined with PIWI subfamily member Piwi protein or AGO3 protein to play a role.
- Small-molecule interfering RNA (siRNA): siRNA is a long-chain linear dsRNA, which is processed by Dicer into mature siRNA with a length of 20-24 nt, and is loaded into RISC to guide silencing. Similar to miRNAs, they mediate post-transcriptional silencing through a process called RNA interference (RNAi), in which siRNA interferes with the expression of complementary nucleotide sequences.
- Long non-coding RNA (lncRNA): IncRNA is a non-coding RNA with a length greater than 200 nucleotides. Studies have shown that IncRNA plays an important role in many life activities such as dose compensation effect, epigenetic regulation, cell cycle regulation and cell differentiation regulation, and is a research hotspot in genetics.
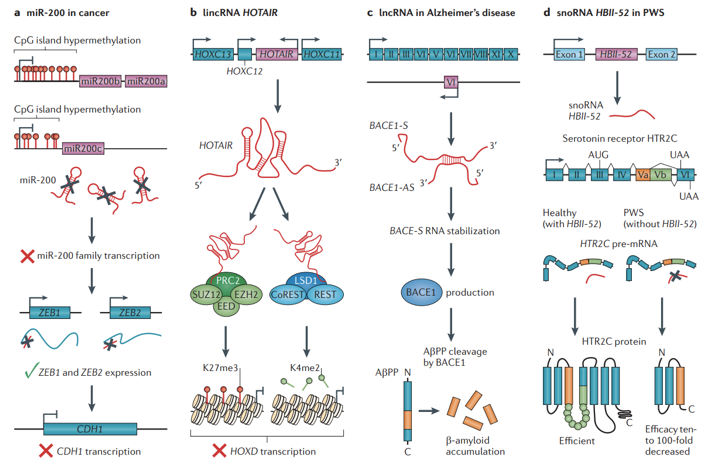 Fig 2. Examples of roles of non-coding RNAs in disease pathophysiology. (Esteller M. 2011)
Fig 2. Examples of roles of non-coding RNAs in disease pathophysiology. (Esteller M. 2011)
What We Offer
Biomedical-Bioinformatics, a division of CD Genomics, provides analysis of different types of regulatory ncRNAs (such as miRNA, piRNA, siRNA, lncRNA, and circular RNA) to explore the regulatory functions of ncRNA in the process of gene expression. For different non-coding RNA data, we provide different software or packages for data analysis according to the properties and characteristics of different non-coding RNAs. In addition to analyzing the regulation of a single ncRNA to its gene or protein, we also provide analysis of the mutual regulation between a variety of ncRNAs to fully meet your data analysis needs. You only need to provide us with your original data or other intermediate data files. We will evaluate the data, preprocess it, perform non-coding RNA regulation analysis, and finally generate a complete report, in which high-quality and easy-to-interpret images and tables can be used for publication of the article.
Data Ready
Before data analysis, the first thing is to get your data ready. For non-coding RNA regulation analysis, the raw input data can be experimental raw data generated in your own laboratory or that of a 3rd party provider or downloaded from public databases.

For non-coding RNA regulation analysis services, if you have any questions about the data analysis cycle, analysis content and price, please click online inquiry.
What's More
For non-coding RNA regulation analysis, if you have yet to generate the raw data, CD Genomics relies on years of experience in ncRNA sequencing services to provide you with different types of ncRNA sequencing services. In addition, we also provide services to obtain raw data from public databases. In short, we are happy to work with you at every stage of your research to ensure the best outcome for your study. If you have any questions about our non-coding RNA regulation analysis, please feel free to contact us for details.
References
- Esteller M. Non-coding RNAs in human disease[J]. Nat Rev Genet. 2011 Nov 18;12(12):861-74. doi: 10.1038/nrg3074. PMID: 22094949.
- Mattick JS, Makunin IV. Non-coding RNA[J]. Hum Mol Genet. 2006 Apr 15;15 Spec No 1:R17-29. doi: 10.1093/hmg/ddl046. PMID: 16651366.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
