Whole Genome Multilocus Sequence Typing
Introduction
Multilocus sequence typing (MLST) is a bacterial typing method based on nucleic acid sequence determination. This method uses PCR to amplify multiple internal fragments of housekeeping genes and determine their sequences to analyze the variation of strains. MLST is simple to operate, results can be obtained quickly and are convenient for comparison between different laboratories, and has been used for epidemiological monitoring and evolutionary research of various bacteria. With the acceleration of sequencing, the reduction of costs, and the development of analysis software, MLST has gradually become a routine method for bacterial typing. As the second-generation sequencing technology provides a fast, cost-effective and efficient method for bacterial genome sequencing, as well as the development of analysis software, the whole genome multisite sequence typing (wgMLST) technology has gradually replaced the traditional MLST Sanger sequencing technology. More loci are considered in wgMLST typing analysis (usually 1500-4000), so higher typing resolution can be obtained. Genome-wide multilocus sequence typing is used to study bacterial epidemiology, population biology, pathogenicity and evolution.
The wgMLST method based on the second-generation sequencing data can be applied to various fields:
- Reveal the relationship between bacterial virulence and genotype in epidemiological studies.
- Trace the source and spread of resistant strains.
- Through the analysis of housekeeping gene sequence characteristics, the epidemiological and evolutionary characteristics of a large number of representative strains are studied to provide a basis for preventing the spread of pathogenic bacteria.
Data Analysis Technical Route
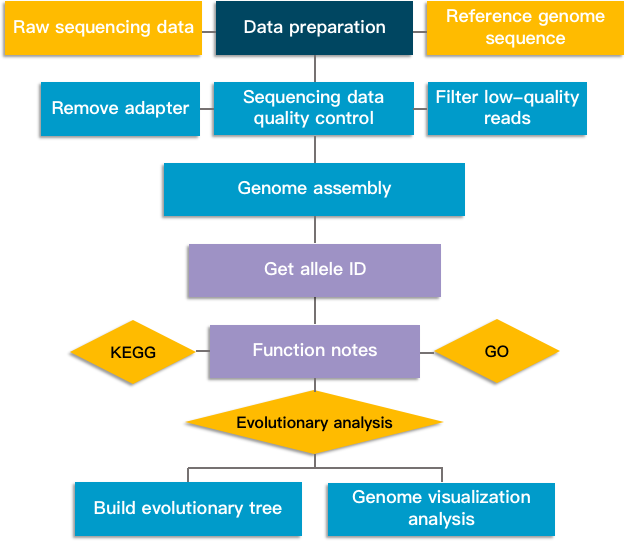 Fig 2. Whole genome multilocus sequence typing analysis pipeline.
Fig 2. Whole genome multilocus sequence typing analysis pipeline.
An Example of wgMLST Analysis Content
1. Original off-machine data processing: This step filters reads with low sequencing quality values, retains high-quality reads, and performs data quality control.
2. Sample assembly: Use Velvet algorithm to assemble the genome, obtain the sequence file that can reflect the basic situation of the sample genome, and evaluate the assembly result.
3. Allele ID acquisition: Based on assembly based and assembly free algorithms, allele ID numbers are acquired for gene loci in the genome.
4. Functional annotation: Perform functional annotation and pathogenicity and drug resistance analysis of different databases for coding gene sequences.
5. Evolutionary analysis: Use wgMLST data to construct high-resolution species typing results, explain the outbreak of infectious diseases, analyze the population structure and evolutionary laws of target bacterial species, and provide a data basis for disease prevention and control.
6. Genome visualization analysis: Combining the results of genome component analysis and functional annotation results to display the basic research results of the genome.
What We Offer
As one of the global biological information analysis service providers, CD Genomics provides established, cost-efficient, and rapid turnaround analysis services for whole genome multilocus sequence typing analysis for researchers, aiming to help you study the microbial molecular typing of pathogenic bacteria and pathogens. CD Genomics provides different cut-off software to perform whole genome multilocus sequence typing analysis for customers to meet the analysis needs of different research directions. In addition, we are able to receive various formats of data for analysis such as raw data files, or other intermediate data formats. You only need to provide us with your original data, and we will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis result report.
Our workflow

What's more, if you do not currently have the genome sequence input data, CD Genomics can also provide you with a variety of sequencing services based on its rich sequencing experience. If you have any questions about the data analysis cycle, analysis content and price, please click online inquiry. With years of experience in biological data analysis, CD Genomics' analysis engineers will provide you with the most appropriate analysis strategy based on your data, and generate high-quality results and charts that can be used for publication. For more detailed information, such as sequencing services and other data analysis services, please feel free to contact us.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
