Whole Exome Genotyping Analysis
Introduction
Whole exome sequencing is a widely used next-generation sequencing (NGS) method that involves the sequencing of protein coding regions of the genome. The human exome accounts for no more than 2% of the genome, but it contains about 85% of the known disease-related variants, which makes this method a cost-effective alternative to whole-genome sequencing. Exome sequencing can detect mutations in coding exons, expand the target content, including untranslated regions (UTR) and microRNA, and obtain a comprehensive view of gene regulation. When whole-genome sequencing is impossible or unnecessary, exome sequencing is a cost-effective alternative. Sequencing only the coding regions of the genome allows researchers to focus their resources on the genes most likely to affect the phenotype, with shorter turnaround times and lower prices. Analysis of exome sequencing data, such as typing analysis, can effectively identify coding variants in a wide range of applications including population genetics, genetic disease and cancer research.
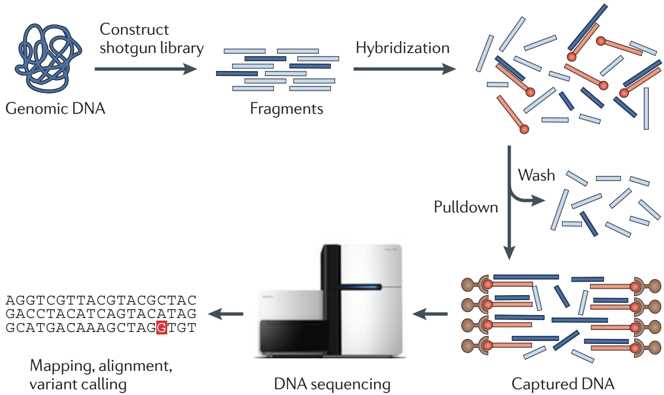 Fig 1. Workflow for exome sequencing. (Bamshad MJ, et al. 2011)
Fig 1. Workflow for exome sequencing. (Bamshad MJ, et al. 2011)
Application Areas of Whole Exome Genotyping Analysis
Whole exome sequencing (WES) is a sequencing technology for exons with protein coding functions. In recent years, the use of whole exome sequencing for genotyping has gradually been widely used in the discovery of exon gene mutations.
- Exome sequencing has deeper coverage and higher data accuracy, the cost of genome resequencing is lower, and it has greater advantages for studying disease-related SNPs and Indels.
- It has the function of mutation recognition for a wide range of applications.
- Achieve full coverage of the coding area.
- Provide a cost-effective alternative to whole-genome sequencing, each exome is sequenced 4–5 Gb, and each whole human genome is sequenced about 90 Gb.
- Compared with the whole genome method, the generated data set is smaller and easier to manage, so it can be analyzed more simply and quickly.
Data Analysis Technical Route
The whole exome sequencing data analysis can be used for Mendelian genetic disease research and various complex disease research. The identification of traditional Mendelian disease genes is mainly determined by Sanger sequencing of candidate genes. The determination of candidate genes is mainly through the location mapping methods of some genomic regions, such as karyotype analysis, linkage analysis and so on. However, this method cannot determine whether the disease is caused by a single nucleotide mutation or a variation in genome structure. With the development of second-generation sequencing technology, especially the emergence of exome sequencing technology, common and rare mutations in the genome and protein coding regions of the genome can be accurately detected, which greatly promotes the identification of disease genes. The following flowchart shows the Mendelian genetic disease research program based on whole exome sequencing data analysis:
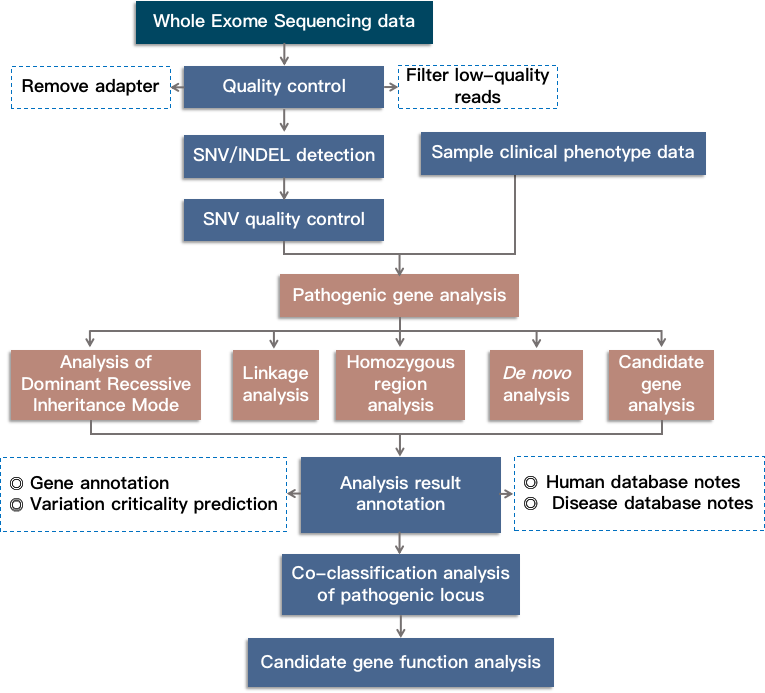 Fig 2. Bioinformatics analysis route for Mendelian genetic disease research based on whole exome sequencing.
Fig 2. Bioinformatics analysis route for Mendelian genetic disease research based on whole exome sequencing.
What We Offer
As one of the global biological information analysis service providers, CD Genomics provides established, cost-efficient, and rapid turnaround analysis services for whole exome genotyping typing analysis for researchers, aiming to help you efficiently identify mutations in the genome. CD Genomics provides different cut-off software to perform whole exnome genotyping typing analysis for customers to meet the analysis needs of different research directions. In addition, we are able to receive various formats of data for analysis such as raw data files, or other intermediate data formats. You only need to provide us with your original data, and we will be responsible for all the follow-up matters of the project, and finally provide you with a complete and easy-to-interpret analysis report.
Advantages of CD Genomics
What’s more
If you do not currently have the exome sequence input data, CD Genomics can also provide you with a variety of sequencing services based on its rich sequencing experience. If you have any questions about the data analysis cycle, analysis content and price, please click online inquiry. With years of experience in biological data analysis, CD Genomics' analysis engineers will provide you with the most appropriate analysis strategy based on your data, and generate high-quality results and charts that can be used for publication. For more detailed information, such as sequencing services and other data analysis services, please feel free to contact us.
References
- Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery[J]. Nat Rev Genet. 2011 Sep 27;12(11):745-55.
- Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth[J]. Am J Hum Genet.2012 Oct 5;91(4):597-607.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
