COG Enrichment Analysis
Introduction of COG Enrichment Analysis
COG (Clusters of Orthologous Groups) annotation is a method of functional annotation of differential genes. COG is a protein database created and maintained by NCBI. It is a database for homologous classification of gene products, an early database for identifying orthologous genes, and a large number of comparisons of protein sequences from various organisms. COG is divided into two categories, one is prokaryotes and the other is eukaryotes. Prokaryotes are generally called COG databases; eukaryotes are generally called KOG databases. A certain protein sequence can be annotated into a certain COG through alignment, and each cluster of COG is composed of orthologous sequences, so that the function of the sequence can be inferred. COG databases can be divided into 26 categories according to their functions.
Applications of COG Enrichment Analysis in Biology
In biological research, after identifying differentially expressed genes through sequence analysis, genes can be annotated according to their functions (usually based on COG, GO and KEGG databases) to evaluate the effects of different gene expressions on biological functions, and finally find the molecular target. The function of COG annotation: Firstly, the unknown sequence is functionally annotated by known proteins; secondly, by checking the number, presence and absence of the protein corresponding to the specified COG number, it is possible to deduce whether a specific metabolic pathway exists; finally, each COG number represents a class of proteins. Multi-sequence alignment of the query sequence and the COG-numbered proteins on the alignment can identify conserved sites and analyze their evolutionary relationship.
An Example of GOG Enrichment Analysis
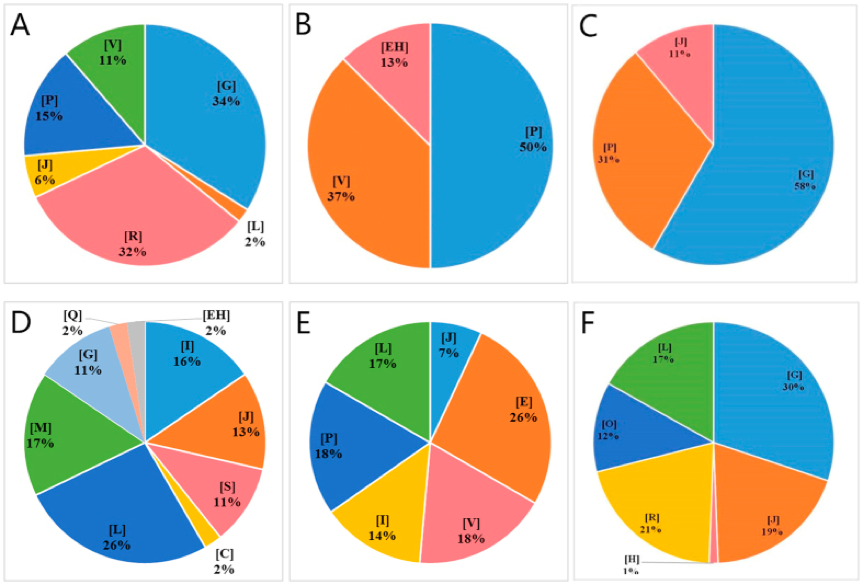 Figure1. Significantly enriched COG terms of differentially expressed genes. (Liu J, et al. 2016)
Figure1. Significantly enriched COG terms of differentially expressed genes. (Liu J, et al. 2016)
- Different colors represent different COG function annotation terms. V: Defense mechanisms; P: Inorganic ion transport and metabolism; J: Translation, ribosomal structure and biogenesis; R: General function prediction only; G: Carbohydrate transport and metabolism; EH: Thiamine pyrophosphate-requiring enzymes; M: Cell wall/membrane/envelope biogenesis; L: Replication, recombination and repair; I: Lipid transport and metabolism; S: Function unknown; E: Amino acid transport and metabolism; H: Coenzyme transport and metabolism; O: Posttranslational modification, protein turnover, chaperones; Q: Secondary metabolites biosynthesis, transport and catabolism; C: Energy production and conversion.
- The area of the pie chart represents the percentage of the number of differentially expressed genes annotated to this COG term.
What We Offer
CD Genomics provides different types of gene function annotation analysis services. For COG annotation and enrichment analysis, in addition to providing pie charts, we also provide bar charts and other intuitive display methods (such as circle charts that integrate multiple information). We provide high-quality COG annotation enrichment analysis pie chart or histogram, which allows you to quickly understand the functions of related proteins and meet your needs for publishing articles. Different software or analysis tools are used for COG enrichment analysis to meet your personalized analysis needs. CD Genomics provides researchers with one-stop, mature, cost-effective and fast turnaround analysis services to help researchers mine the function of differentially expressed genes in different samples.
Data Ready
Before COG enrichment analysis, the first thing is to get your data ready. We can use different types of sequencing data or other experimental data, and data files can be raw data or intermediate data formats (such as differential gene expression files). The raw data or intermediate data can be obtained from the following channels:
If you don't have the data for COG annotation and enrichment analysis, CD Genomics can also provide you with different types of sequencing services or download related data from existing open databases. If you have any questions about the data analysis content, turnaround time and price, please click online inquiry.
Our Service Process
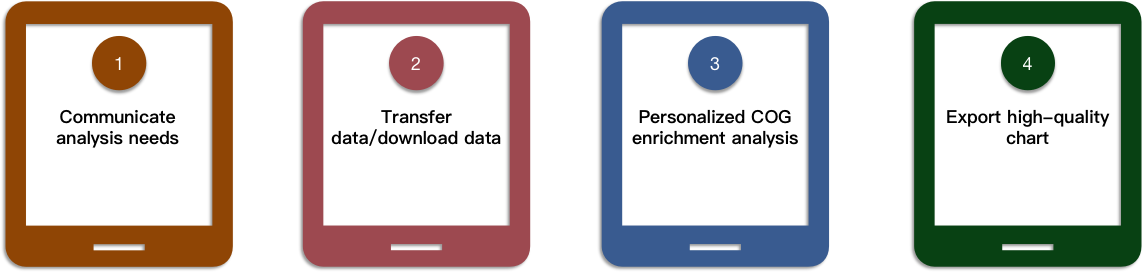
Biomedical-Bioinformatics, a division of CD Genomics, provides COG annotation and enrichment analysis service according to customer's requirements. With years of data analysis experience, CD Genomics provides you with high-quality gene annotation analysis services and provides a reliable data basis for your wet experiments. In addition to COG enrichment analysis, we also provide various types of gene function annotation analysis services, such as KEGG pathway enrichment analysis and GO enrichment analysis. For COG enrichment analysis, if you have any questions, please feel free to contact us. We have a professional technical support team to provide you with the best services, and we look forward to working with you!
Reference
- Liu J, et al. Transcriptomic analysis on the formation of the viable putative non-culturable state of beer-spoilage[J]. Scientific Reports, 2016, 6: 36753.
* For research use only. Not for use in clinical diagnosis or treatment of humans or animals.
Online Inquiry
Please submit a detailed description of your project. Our industry-leading scientists will review the information provided as soon as possible. You can also send emails directly to for inquiries.
