Unraveling Wheat Genomes: From Sequencing to Functional Insights
As one of the three staple foods in the world, the annual output of wheat accounts for about 30% of the total grain output in the world, and its stable supply is directly related to the nutritional security of billions of people. However, the complexity and high repeatability of the allohexaploid genome of common wheat seriously hinder the process of molecular breeding and gene function analysis.
With the innovation of the next generation sequencing and chromosome conformation capture technology, mapping the whole genome of wheat has become the key to break through the bottleneck of improving yield, quality and stress resistance, which is of strategic significance to ensure global food security.
This paper introduces the characteristics of wheat genome, expounds the important significance of genome sequencing in agricultural development, reviews the transformation of sequencing technology, and looks forward to its prospects.
Main Characteristic of Wheat Genome
The genome of wheat has unique and complex characteristics, which not only profoundly affect the growth and development, physiological characteristics and environmental adaptability of wheat, but also are closely related to human food security and sustainable agricultural development.
Complex Genome Structure
The wheat genome is extremely huge. The genome size of ordinary wheat is about 16 Gb, which is about five times that of human genome. This huge genome scale is mainly attributed to the existence of a large number of repetitive sequences, among which transposable elements (TE) are the most prominent components. Transposons can move and replicate in the genome, resulting in its proportion in the wheat genome as high as 80%. The existence of these repetitive sequences makes the assembly of wheat genome face great challenges, and also provides a rich genetic material basis for the evolution of genome and the generation of new functions.
In addition, wheat is an allohexaploid (AABBDD), which is formed by two natural hybridization and polyploidization events from three different diploid ancestor species (urartu wheat A genome, Aegilops spelt B genome and Aegilops tauschii D genome). This polyploid characteristic leads to three sets of highly similar but not identical subgenomes in wheat genome.
There are a large number of homologous genes between subgenomes, that is, partial homologous genes, which have high similarity in sequence, but there may be differences in expression patterns and functions. This complex polyploid structure endows wheat with rich genetic diversity, which enables it to survive and reproduce in different ecological environments, but also brings challenges to gene function research and genetic breeding.
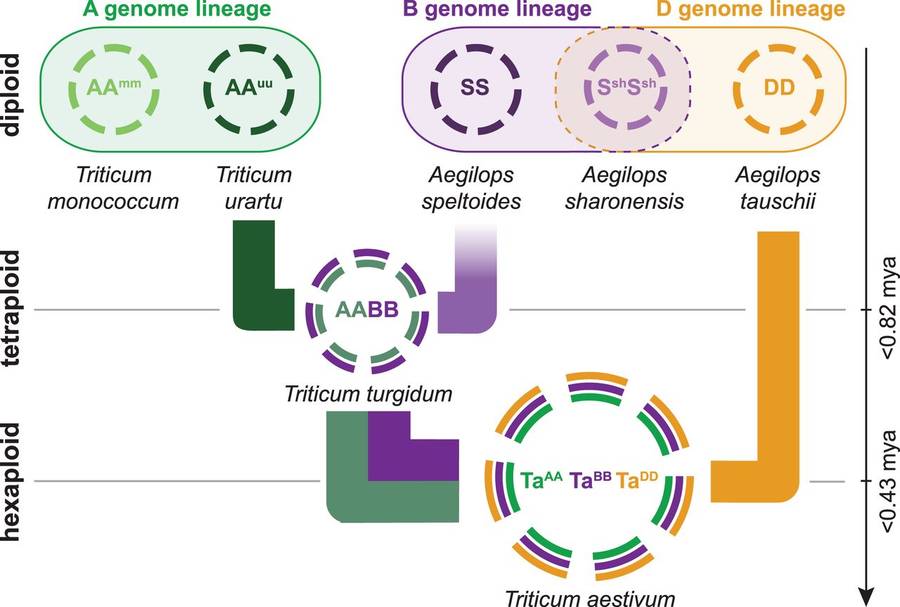 Schematic diagram of the relationships between wheat genomes with polyploidization history and genealogy (IWGSC., 2014)
Schematic diagram of the relationships between wheat genomes with polyploidization history and genealogy (IWGSC., 2014)
Functional Diversity of Genes
The wheat genome encodes a large number of genes, which is estimated to be around 100,000, far exceeding many other plant species. These genes are widely distributed on various chromosomes, forming numerous gene families. The expansion and contraction of gene family is one of the important driving forces of wheat genome evolution. The NBS-LRR (Nucleotide-Binding Site-Leucine-Rich Repeat) gene family related to disease resistance has expanded significantly in the wheat genome, with hundreds of members. Different NBS-LRR genes have specific recognition ability for different pathogens, which enables wheat to cope with complex and changeable pathogen environment.
In addition, the wheat genome is also rich in gene families related to stress resistance (such as drought, salinity and low temperature). Dehydrin gene family is expressed in large quantities under drought stress. By combining with ions and biological macromolecules in cells, the osmotic pressure and membrane stability of cells are maintained, thus enhancing the drought tolerance of wheat. These rich gene families and the synergy between their members make wheat have a strong ability to adapt to different environmental conditions.
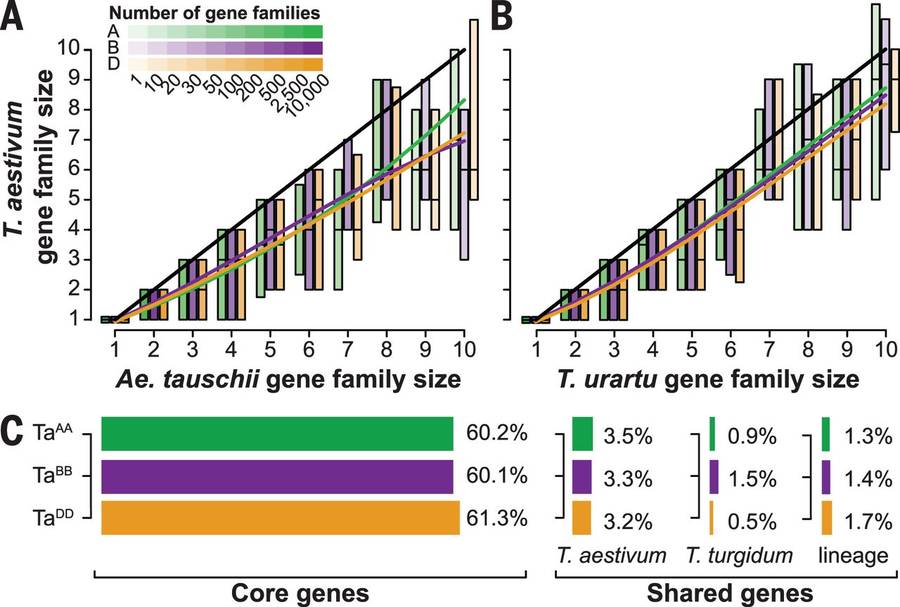 Gene conservation and the wheat pan- and core genes (IWGSC., 2014)
Gene conservation and the wheat pan- and core genes (IWGSC., 2014)
Extensive Gene Expression Regulation Network
The complexity of wheat genome is not only reflected in structure, but also in gene expression regulation. Due to polyploid characteristics and the existence of a large number of repetitive sequences, the wheat genome has formed a complex gene expression regulation network. Among them, epigenetic regulation plays an important role in wheat gene expression regulation. DNA methylation, histone modification and non-coding RNA (such as miRNA, lncRNA, etc.) mediate the regulation mechanism to regulate the gene expression level.
In addition, transcription factors also play a central role in the regulation network of wheat gene expression. Different types of transcription factors (such as MYB, bHLH, AP2/ERF, etc.) activate or inhibit the transcription of downstream genes by binding to cis-acting elements in gene promoter region. These transcription factors interact with each other to form a complex regulatory network, which accurately regulates the gene expression patterns of wheat in different growth and development stages and environmental conditions, thus achieving fine regulation of various biological processes.
Services you may interested in
Bulked Segregant Analysis (BSA) Services
Why is Wheat Genome Sequencing Important
As a major breakthrough in the field of modern biology sciences, wheat genome sequencing is far more significant than the simple analysis of gene information, and it is providing powerful help to solve the food challenge faced by human beings and promote the sustainable development of agriculture.
Genetic Improvement
Wheat genome sequencing is of great significance to wheat genetic improvement. The wheat genome is huge and complex, and the proportion of repeated sequences is extremely high, which makes the traditional breeding work more difficult. Through genome sequencing, researchers can accurately locate genes related to excellent agronomic traits, such as high-yield genes and high-quality protein genes. In addition, after understanding the gene function, breeders can also use advanced technologies such as molecular marker-assisted breeding and gene editing to break the barriers of gene communication among species, quickly aggregate multiple excellent genes, and cultivate new wheat varieties with high yield, high quality and stress resistance, which can significantly shorten the breeding cycle and improve the breeding efficiency.
Wheat genome sequencing is also a key means to cope with environmental changes. In recent years, global climate change has intensified and extreme weather has occurred frequently. Drought, high temperature, floods and other disasters pose a serious threat to wheat production. Different wheat varieties have different response mechanisms to environmental stress, and genome sequencing can reveal the genetic basis behind these differences. For example, by analyzing the genome of drought-tolerant wheat varieties, scientists have discovered a series of genes and regulatory pathways related to drought tolerance.
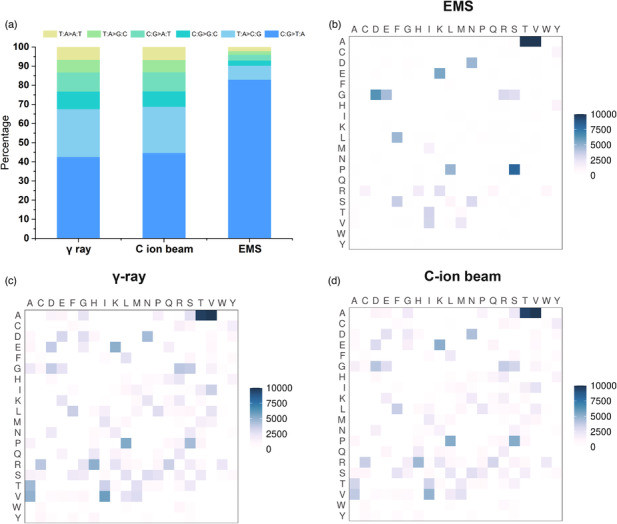 Comparison of the percentage of different types of SNP variations and the amino acid changes induced by EMS, γ-ray, or C-ion beam irradiation in the mutant population (Xiong et al., 2023)
Comparison of the percentage of different types of SNP variations and the amino acid changes induced by EMS, γ-ray, or C-ion beam irradiation in the mutant population (Xiong et al., 2023)
Encourage Scientific Research
From the perspective of scientific research, wheat genome sequencing has greatly enriched the research content of plant genomics. As an important representative of Gramineae, wheat genome has certain homology and evolutionary relationship with rice, corn and other crops. The in-depth study of wheat genome is helpful to reveal the evolution law of plant genome, the origin and evolution of gene family, and provide key clues for understanding the basic mechanism of plant life activities. At the same time, wheat genome sequencing has also promoted the development of comparative genomics, functional genomics and other related disciplines, and provided important theoretical basis and technical support for the research in the whole field of plant science.
Promote Agricultural Development
Wheat genome sequencing is also of great value in the development of agricultural industry. The cultivation of high-quality special wheat varieties can meet the diversified needs of food processing industry, such as strong gluten wheat for making bread and weak gluten wheat for making biscuits. Through genome sequencing technology, breeders can improve wheat quality, improve its processing performance, enhance the added value of agricultural products, and promote the extension and upgrading of agricultural industrial chain.
In addition, molecular markers and gene detection technologies based on genome information can be applied to the fields of wheat seed purity identification, variety authenticity detection, etc., to standardize the seed market promote the healthy and sustainable development of wheat industry.
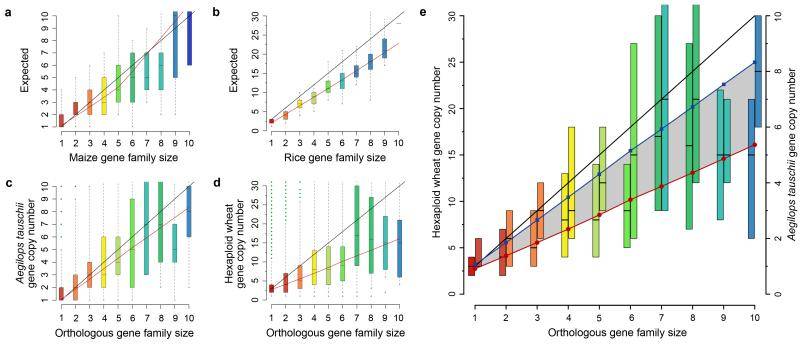 Gene family sizes in orthologous assemblies of hexaploid wheat, Ae. tauschii, simulated maize and hexaploid rice (Brenchley et al., 2012)
Gene family sizes in orthologous assemblies of hexaploid wheat, Ae. tauschii, simulated maize and hexaploid rice (Brenchley et al., 2012)
How Has Wheat Genome Sequencing Changed
With the development of genomics and the continuous innovation of sequencing technology, it has brought dawn to overcome the wheat genome problem. The research on the development of wheat genome sequencing technology is helpful to fully understand the mystery of wheat genome, promote wheat genetic improvement and molecular breeding, and ensure global food security.
Traditional Sanger Sequencing Method
In the early stage of genome sequencing, Sanger sequencing is the main technical means. Sanger sequencing method is based on the principle of dideoxynucleotide termination, which reads the base sequence by synthesizing a strand complementary to the template DNA. This method has the advantage of high accuracy and can obtain high-quality sequence data. In the early stage of wheat genome research, scientists tried to sequence some gene fragments of wheat by Sanger sequencing method.
Because of the huge genome of wheat, it is expensive and the throughput is extremely low to use Sanger sequencing method to sequence the whole genome. It takes a lot of manpower, material resources and time to complete the whole genome sequencing of wheat, which is almost impossible under the technical and resource conditions at that time. Nevertheless, Sanger sequencing method laid a foundation for the follow-up study of wheat genome, and made researchers have a preliminary understanding of the local characteristics of wheat genome.
Construction of Physical Map Based on BAC Library
In order to overcome the limitations of Sanger sequencing method in wheat genome sequencing, scientists adopted the strategy of physical map construction based on bacterial artificial chromosome (BAC) library. Firstly, the BAC library of wheat genome is constructed, that is, the DNA fragments of wheat genome are inserted into BAC vector, and Escherichia coli is transformed to form clones containing different wheat genome fragments. Then, these clones were sorted and screened to construct physical maps. By sequencing BAC clones, the sequence of wheat genome can be spliced step by step.
The advantage of this method is that it can decompose the huge and complex wheat genome into relatively small and easy-to-operate fragments for research. This method has the problems of complicated operation and huge workload. It requires a lot of experimental operations and data analysis to construct physical maps, and it is also difficult to splice different BAC clones, which is prone to errors and gaps.
New Generation Sequencing Technology
The emergence of Next Generation Sequencing (NGS) has brought revolutionary changes to wheat genome sequencing. Compared with the traditional sequencing technology, the new generation sequencing technology has obvious advantages of Qualcomm and low cost. It can simultaneously sequence a large number of DNA fragments in a short time, which greatly improves the sequencing efficiency. Moreover, with the development and large-scale application of technology, the sequencing cost has been greatly reduced, which makes it possible to sequence such a huge genome as wheat. The new generation sequencing technology can also detect some genetic variations that are difficult to be found by traditional techniques, such as single nucleotide polymorphism (SNP) and insertion deletion (InDel).
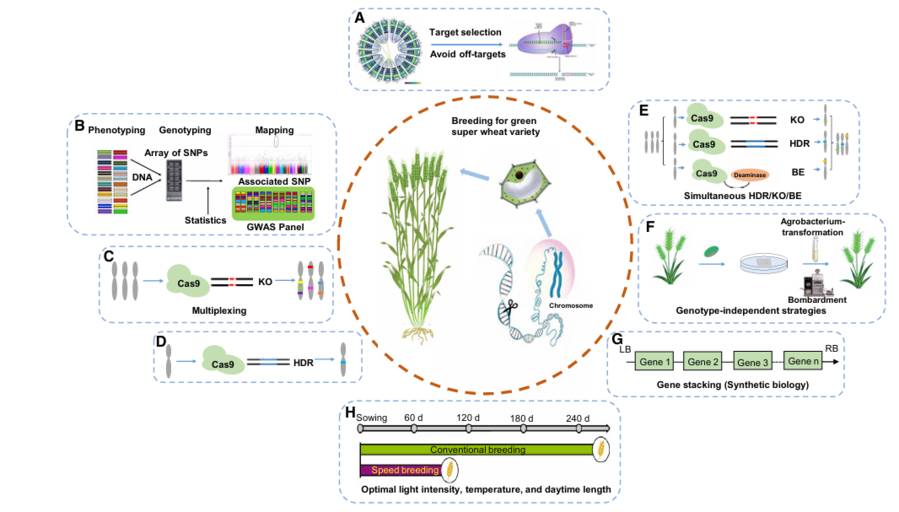 Breeding of a green super wheat variety through CRISPR/Cas-mediated gene editing and other breeding technologies (Li et al., 2021)
Breeding of a green super wheat variety through CRISPR/Cas-mediated gene editing and other breeding technologies (Li et al., 2021)
Future Prospects of Wheat Genome Sequencing
With the innovation of technology and the deep integration of multi-disciplines, wheat genome sequencing is moving towards a new stage of development.
Promote Precision Breeding
With the continuous progress and perfection of wheat genome sequencing technology, the future breeding work will move towards unprecedented accuracy. Scientists can accurately locate genes closely related to key traits such as yield, quality, disease resistance and stress resistance according to a complete and accurate genome map, just like operating precision instruments.
In breeding practice, breeders can use these genetic information to select suitable parents for hybridization, which greatly improves the probability and efficiency of cultivating new wheat varieties with high yield, high quality and strong stress resistance, shortens the breeding cycle, accelerates the popularization and application of new varieties, and lays a solid foundation for the stability and security of global food supply.
Respond to Complex Environmental Challenges
The frequent occurrence of extreme weather caused by global climate change has brought severe tests to wheat production. With the help of wheat genome sequencing results, researchers can deeply explore the molecular mechanism of wheat responding to environmental stress. When studying how to resist drought in wheat, the key gene switch to start the drought-resistant defense mechanism was found by analyzing the expression changes of related genes in the genome.
It is introduced into cultivated wheat by modern biotechnology to enrich the genetic diversity of wheat, cultivate wheat varieties adapted to different complex environments, and ensure that wheat can achieve stable and high yield under various unfavorable conditions.
Expand Wheat Germplasm Resources
At present, there are still a large number of potential excellent genes in the known wheat germplasm resources that have not been fully tapped and utilized. With the wide application of genome sequencing technology in more wheat varieties (including ancient local varieties and wild relatives), a more comprehensive and detailed genome variation map of wheat can be constructed. Through the systematic analysis of genomes of different germplasm resources, we can find some genes that have been lost or rare in modern cultivated varieties, and these genes may contain important characters for future wheat breeding, such as unique disease and pest resistance mechanism and special quality characteristics.
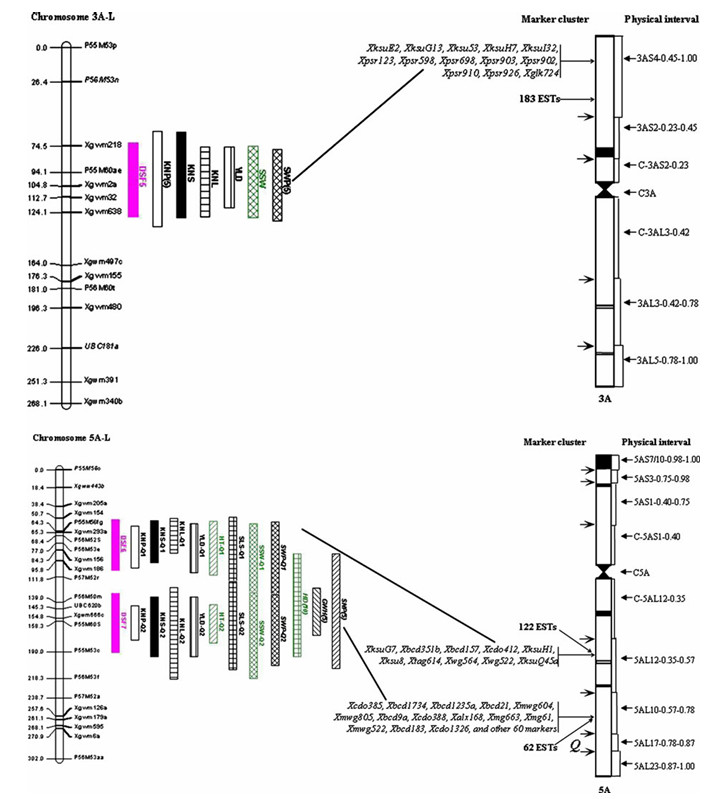 Map and physical locations of DSFs and their involved QTLs in L version maps of wild emmer wheat T. dicoccoides (Peng et al., 2011)
Map and physical locations of DSFs and their involved QTLs in L version maps of wild emmer wheat T. dicoccoides (Peng et al., 2011)
Conclusion
The completion of wheat genome sequencing has brought a breakthrough in plant genomics research and innovated the model of crop genetic improvement. Combined with cutting-edge technology, research can accurately locate key genes. In the future, genomics will drive the development of molecular breeding and gene editing technology, help cultivate high-quality wheat varieties and support the sustainable development of agriculture.
References
- International Wheat Genome Sequencing Consortium (IWGSC). "A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome." Science. 2014 345(6194):1251788 https://doi.org/10.1126/science.1251788
- Xiong H, Guo H., et al. "A large-scale whole-exome sequencing mutant resource for functional genomics in wheat." Plant Biotechnol J. 2023 21(10):2047-2056 https://doi.org/10.1111/pbi.14111
- Brenchley R, Spannagl M., et al. "Analysis of the bread wheat genome using whole-genome shotgun sequencing." Nature. 2012 491(7426):705-10 https://doi.org/10.1038/nature11650
- Shaoya Li, Chen Zhang., et al. "Present and future prospects for wheat improvement through genome editing and advanced technologies" Plant Communications 2021(2): 100211 https://doi.org/10.1016/j.xplc.2021.100211
- Peng, J.H, Sun D., et al. "Domestication evolution, genetics and genomics in wheat." Mol Breeding 2011(28): 281–301 https://doi.org/10.1007/s11032-011-9608-4
 Send a Message
Send a MessageFor any general inquiries, please fill out the form below.