Key Databases for Wheat Research: An Overview of Features and Applications
Bread wheat is one of the earliest domesticated crops and has been the staple food of human beings for thousands of years. Wheat is now the most widely planted crop in the world, and its trade value exceeds the sum of all other crops. Since 2013, the genomes of wheat ancestors (AA) and Aegilops tauschii (DD) and the genomes of potential candidates for B genome ancestors have been assembled. Since then, wheat genomes with different ploidy levels have been assembled and released, including the genomes of wild emmer, durum and several hexaploid wheat varieties.
In order to access and use these data conveniently and effectively, researchers have developed several wheat genomics databases in recent years. Through these databases, we can quickly obtain wheat genome and other related data, including genome-wide sequencing (WGS), transcriptomics, epigenomics and protein genomics data. Some of these databases integrate a series of useful tools, such as WheatOmics for expression and co-expression analysis, BSA analysis module based on gene map in WheatGmap and function prediction process based on genome-wide functional gene network in WheatNet.
Services you may interested in
Bulked Segregant Analysis (BSA) Services
Comprehensive Database
Wheat Genome Information Database (http://www.wheatgenome.org/) International Wheat Genome Sequencing Alliance (IWGSC) is an international cooperative alliance, which was established in 2005 by a group of wheat growers, plant scientists and public and private breeders. The vision of IWGSC is the high-quality genome sequence of bread wheat, which serves as the basis for accelerating the development of improved varieties and enhancing all aspects of basic and applied wheat science.
 Comprehensive Genome Platform for Wheat Gene Mapping and Data Sharing (https://www.wheatgmap.org/) This platform integrates a variety of BSA-based mapping models and a large number of public data, which helps researchers to use BSA method for wheat gene cloning and functional research, while managing and sharing sequencing data and phenotypic data.
Comprehensive Genome Platform for Wheat Gene Mapping and Data Sharing (https://www.wheatgmap.org/) This platform integrates a variety of BSA-based mapping models and a large number of public data, which helps researchers to use BSA method for wheat gene cloning and functional research, while managing and sharing sequencing data and phenotypic data.

GrainGenes, the molecular and phenotypic information database of Triticum and Oats (http://wheat.pw.usda.gov), is a comprehensive resource dedicated to providing molecular and phenotypic information about wheat, barley, rye and other related crops (including oats). This resource brings together a wide range of genetic data and research results, providing a convenient and comprehensive information exchange platform for researchers. Through GrainGenes, researchers can easily obtain genetic information, gene sequence, expression data and phenotypic data of crops, thus promoting the research progress in the field of crop genetics and genomics.
Functional Genome Database
Online platform for wheat genomics data (http://wheatomics.sdau.edu.cn/) This platform integrates and visualizes 48 sets of wheat genome, more than 2,000 wheat variation groups, 3 sets of wheat mutant library exon groups, more than 500 wheat transcription groups and several sets of wheat apparent modification groups. On this basis, functional modules such as gene identification, gene expression/co-expression analysis, gene function analysis, interaction protein identification and epigenetics analysis were developed, which provided convenient tools for wheat functional genome research and solved the problem that wheat researchers at home and abroad could not obtain target genes and data quickly and effectively.

Wheat Homologous Gene Database (http://wheat. cau. edu.cn/TGT/) This database contains a wide range of gene sequences of wheat plants, including the identification, comparison and analysis results of homologous genes, in order to promote the research progress in the field of wheat plant genetics and genomics. Through this database, researchers can obtain the needed genetic data more conveniently and conduct in-depth analysis and discussion, thus providing strong support for the breeding, improvement and application of wheat plants.
Interactive platform for exploring functional genes in wheat by integrated gene regulatory network (http://wheat.cau.edu.cn/wGRN/) WGRN is a free interactive platform, which uses integrated gene regulatory network in wheat to guide the discovery of functional genes. This platform brings together the target regulation of transcription factor (TF) from large-scale functional data sets, and provides a series of general analysis tools for research to mine functional genes and regulation for crop improvement. All analyses are based on the genome of IWGSC v2.1 and updated annotations.
Genetic Variation Breeding Database
Genome variation data set of wheat and its ancestors (http://wheat.cau.edu.cn/Wheat_SnpHub_Portal/) This database contains six sets of published variation data of wheat and its ancestors, which provides valuable resources for functional genomics research, genetic breeding and adaptive evolution research of wheat. Through in-depth analysis of these variation data, researchers can reveal the important genetic characteristics of wheat and provide scientific basis for wheat improvement and cultivation.
The database of wheat genome variation and selection signals (https://db.cngb.org/WGVD/) includes genome variation of whole genome resequencing and exon capture data of bread wheat and its ancestors, as well as genome-wide selection signals during wheat domestication and improvement, which promotes the research of wheat function and breeding. SNPs and indels collected from 968 bread wheat and their ancestors, as well as the selection characteristics in the process of wheat domestication and improvement based on SNPs evaluation of 93 wheat genome-wide retest data were summarized in the database. In the current version, WGVD includes 7,346,814 SNPs and 1,044,400 indel, focusing on gene regions and upstream/downstream regions.
Wheat WheatCNVb Database (http://wheat.cau.edu.cn/WheatCNVb/) Database Based on the new molecular marker system of wheat CNVb, the first CNVb digital fingerprint of wheat germplasm in the world was constructed. Combined with ultra-low depth sequencing, CNVb fingerprinting realized the accurate identification of germplasm resources with low cost, Qualcomm amount and high resolution. CNVb marker supports the accurate identification of important breeding utilization alleles, and provides a low-cost, Qualcomm-efficient molecular marker for wheat design and breeding. The new fingerprinting technology is expected to provide strong support for digital management of wheat breeding resources and accurate breeding decision.
WheatCompDB (http://wheat.cau.edu.cn/WheatCompDB/), a comparative database of blood relationship intervals of wheat germplasm resources, provides a comparative analysis of blood relationship intervals of germplasm resources between any two samples in the data set, and supports the dynamic construction and mining of germplasm resources networks at the level of whole genome, single chromosome and local interval.
Protein Omics Database
Wheat protein Group Database (https://wheatproteome. org/) The core of this database is a protein omics study on organs and development stages, which depicts 24 sample types in clickable images on the homepage. Five RNAseq data sets are also provided, with contextual information in the form of annotations and integration with the whole cell metabolic network. As a tool, the database is used to create and retrieve targeted protein group analysis. The "mass westerns" contained in it may enable wheat researchers and breeders to obtain protein-level quantitative data widely, which, combined with the core facilities of mass spectrometry, avoids the need for specific professional knowledge, lengthy development time and low flux related to traditional protein quantitative technology.
Application of Wheat Genome Database
In the field of wheat research and breeding, various professional databases have become an indispensable tool. They cover multi-dimensional data resources such as genome, transcriptome and protein group, and integrate functional modules such as gene location, functional analysis and genetic variation mining. Through these databases, researchers can quickly obtain key data, deeply analyze the genetic mechanism of wheat, accurately locate target genes, and provide solid data support and technical support for cultivating new wheat varieties with high yield, high quality and stress resistance.
Analysis of Genetic Basis of Excellent Wheat Varieties
It is of great significance to analyze the genetic composition of excellent varieties and the law of gene selection and utilization in breeding from the genome level for summing up breeding experience and exploring the genetic basis of important agronomic traits. Taking Aikang 58, an excellent backbone wheat variety, as an example, this paper shows how to use genomics tools to assist in analyzing the genetic composition, structural variation and the breeding and utilization law of important gene/locus haplotypes of wheat varieties.
Bainong Aikang 58 (AK58) is a widely cultivated variety and an important parent of modern breeding in China. Based on the resequencing data, combined with WheatCNVb database, ggComp, IntroBlocker and other tools, this paper analyzed the key parent materials of Aikang 58 in its pedigree from CNV, haplotype and other dimensions. At the same time, haplotype analysis showed that there were centromere-crossing haplotypes (centAHG) differences between the two backbone parents on chromosomes 3B and 6A, while Aikang 58 chose centAHG, the mainstream wheat variety in China, on both chromosomes.
Using the list of known genes compiled by WheatOmics database and SnpHub tool, it was found that ZIM-A1 gene had genotype differences between the two backbone parents. Through the comparative analysis of allelic variation frequency and haplotype network analysis in different evolution stages of wheat, it was found that the haplotype of this gene in Aikang 58 was subjected to selection pressure in the stages of domestication and variety breeding, which proved that Aikang 58 complied with the trend of wheat improvement in haplotype selection of ZIM-A1 gene.
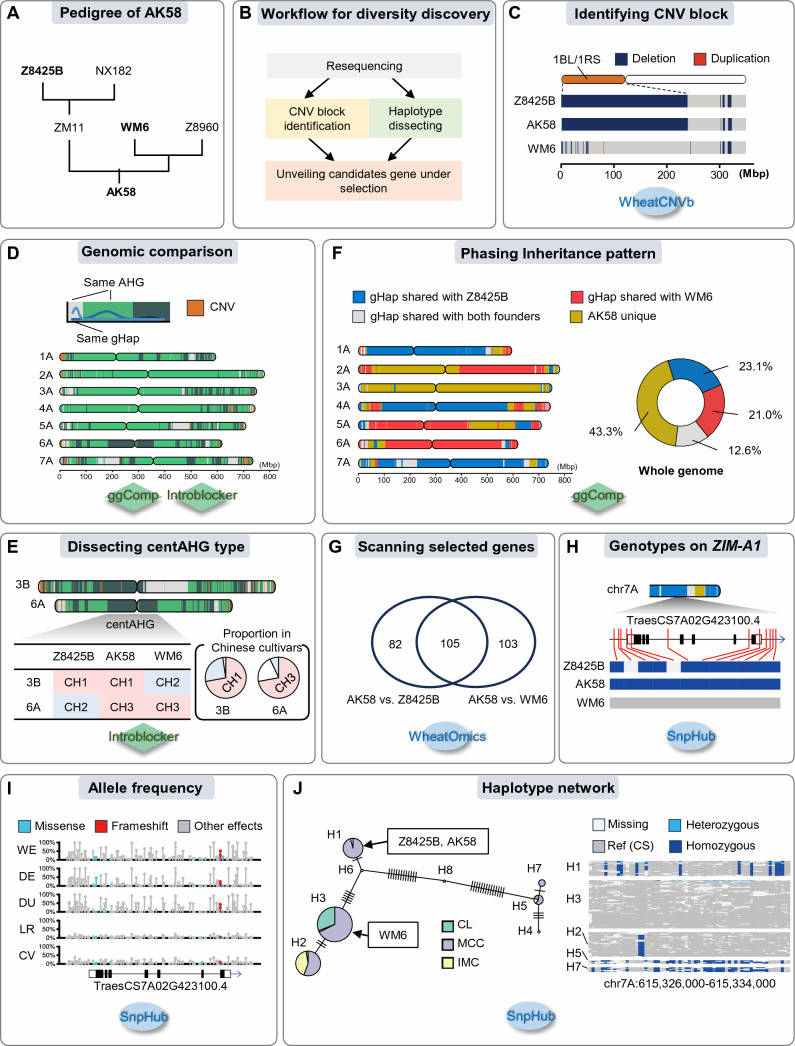 An example of genetic basis analysis of excellent wheat germplasm (Chen et al., 2024)
An example of genetic basis analysis of excellent wheat germplasm (Chen et al., 2024)
Analysis of Functional Gene Exploration-regulation Relationship
Taking drought-resistant gene mining as an example, this paper shows how to use a variety of genomics tools and databases to assist wheat functional gene mining and regulation relationship analysis (Figure 2). Firstly, the published QTLs related to drought stress adaptation were collected, and the drought-resistant candidate genes were prioritized by combining the QTGminer tool of wGRN database and the gene annotation tool of TGT database, and two candidate genes TaNAC071-A and TaWRKY51-1B with high scores were screened out. Then, the gene function prediction of wGRN database, the analysis tool of some homologous genes expression patterns and the gene expression analysis function of WheatOmics database were used to further prove that the two candidate genes may participate in the drought stress response.
Furthermore, the GO enrichment analysis tool of TGT database and the transcription regulatory factor prediction tool of wGRN database were used, and the drought stress response-related regulatory network involved by these two transcription factors was established in combination with the browser of wheat grain translation group, revealing the important role of the two genes in the drought stress response pathway. Using the collinearity analysis tool of TGT database, it was found that the two genes were highly conserved in wheat family species. Combined with the allelic variation frequency analysis of SnpHub database, it was found that both genes were selected during the evolution of wheat.
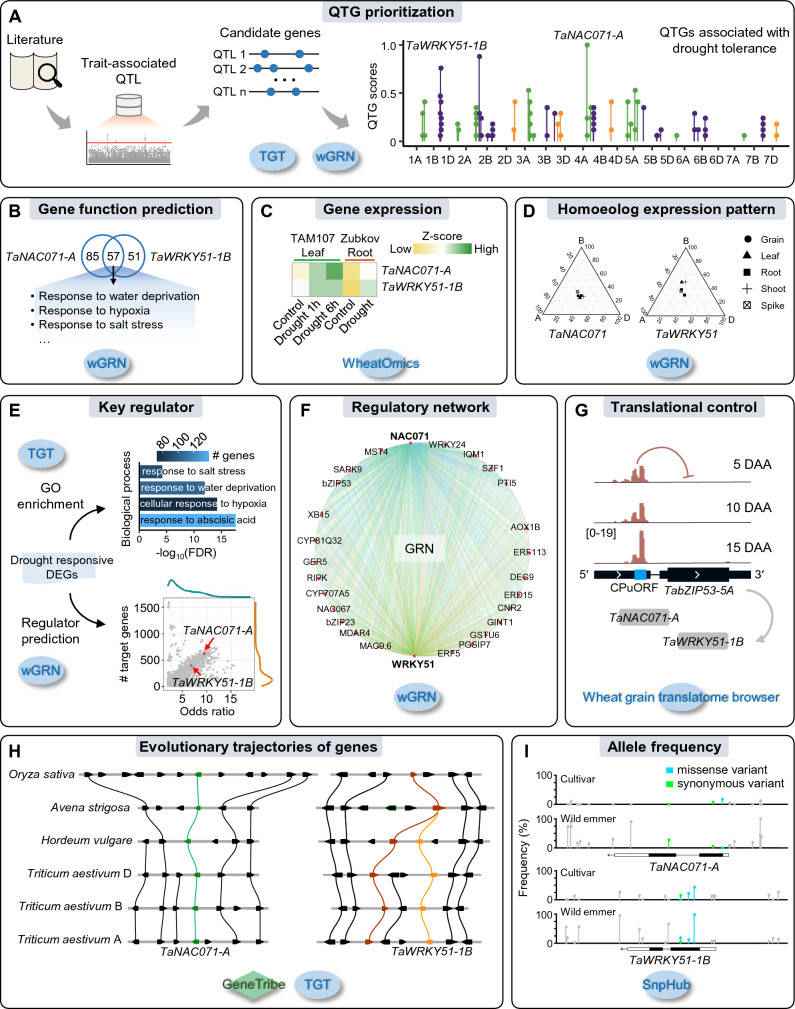 Analysis of the relationship between the exploration and regulation of wheat functional genes (Chen et al., 2024)
Analysis of the relationship between the exploration and regulation of wheat functional genes (Chen et al., 2024)
Conclusion
The above common wheat databases have their own advantages and complement each other, and play a key role in scientific research and breeding practice. From genome sequence analysis to genetic variation mining, from functional gene identification to protein group research, the application of database has greatly accelerated the process of wheat scientific research. In the future, with the continuous updating of data and continuous expansion of functions, these databases will provide stronger support for wheat genetic improvement, variety innovation and industrial development, and help solve major global problems such as food security.
Reference
- Chen Y, Wang W., et al. "Innovative computational tools provide new insights into the polyploid wheat genome." aBIOTECH. 2024 5(1):52-70 https://doi.org/10.1007/s42994-023-00131-7
 Send a Message
Send a MessageFor any general inquiries, please fill out the form below.