2b-RAD Sequencing in Plant Genomics: Protocols, Applications, and Comparative Insights
2b-RAD sequencing technology is a cutting-edge method used in genomics to analyze genetic variations by reducing genome complexity through restriction enzyme digestion and subsequent sequencing. This article provides a comprehensive exploration of the application of 2b-RAD sequencing technology in plant genomics, elaborating on its principles, experimental design, comparisons with other technologies, real-world case studies in plant breeding and agriculture, challenges faced, and proposed solutions, while also offering insights into future development directions. With its unique advantages, 2b-RAD sequencing technology demonstrates immense potential in plant research, serving as a powerful new tool for advancing plant genomics studies and breeding practices.
What is 2b-RAD
In the ongoing research of plant genomics, the demand for efficient and precise whole-genome genotyping technologies has always been crucial. Enter 2b-RAD sequencing technology, which leverages Type IIB restriction enzymes to streamline the whole-genome genotyping process. These enzymes possess unique cutting properties, enabling them to cleave at specific nucleotide sequences and produce uniform fragments. This characteristic allows 2b-RAD technology to analyze plant genomes in a relatively simple and efficient manner, significantly reducing the complexity and cost of whole-genome genotyping.
The RAD-seq (Restriction site Associated DNA sequencing) technology family has continuously evolved, from the early ddRAD (double digest RAD) to the current 2b-RAD, with each iteration aimed at optimizing the workflow for plant research. While ddRAD technology achieved a certain degree of simplified genome sequencing, it still involved some cumbersome steps in the operation. In contrast, 2b-RAD technology builds upon the foundation of ddRAD, further simplifying the operational process and enhancing experimental efficiency through more rational enzyme digestion strategies and experimental designs. This enables researchers to more easily obtain information on plant genomes, providing even stronger technical support for plant genomics research.
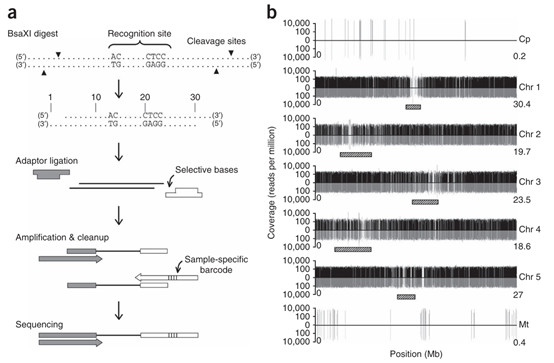 Preparation and sequencing of 2b-RAD tags (Wang et al., 2023)
Preparation and sequencing of 2b-RAD tags (Wang et al., 2023)
Service you may interested in
2b-RAD Principles and Protocol Optimisation
2b-RAD programme design
In the design of 2b-RAD protocols, enzyme selection is a critical step. The BsaXI enzyme demonstrates significant advantages in generating uniform fragment sizes for crops like soybeans. With its specific recognition sequence and cutting sites, BsaXI can evenly cleave DNA in plant genomes, producing fragments of relatively consistent size. This uniform fragment distribution enhances the accuracy and reliability of subsequent sequencing, enabling researchers to obtain genomic information more precisely and laying a solid foundation for downstream gene analysis and breeding applications.
For large-scale agricultural projects, the efficiency of multiplexed sequencing is key to cost-effectiveness. The 2b-RAD technology achieves efficient multiplexed sequencing through optimized experimental design and sequencing strategies. In a single sequencing reaction, multiple samples can be sequenced simultaneously, significantly reducing sequencing costs and time. Additionally, 2b-RAD technology offers flexibility in adapter design. Traditional sequencing techniques often struggle with non-model plants, but 2b-RAD can tailor adapter sequences to the genomic characteristics of non-model plants, thereby improving sequencing success rates and accuracy. In this way, 2b-RAD technology provides an effective means for studying non-model plants, aiding in the exploration of their genetic resources and potential value.
Comparison of ddRAD and 2b-RAD
Compared to ddRAD, 2b-RAD offers significant advantages in terms of operational simplicity. The ddRAD technique requires two enzymatic digestion steps and multiple purification procedures, making the process relatively cumbersome and time-consuming with extensive manual intervention. In contrast, 2b-RAD streamlines the enzymatic digestion and purification steps, reducing manual operation time and enhancing experimental efficiency. This allows researchers to complete experiments in a shorter timeframe, accelerating the research process.
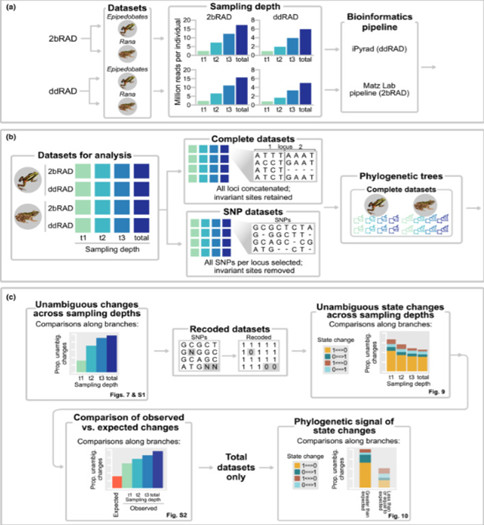 Comparison of ddRAD and 2b-RAD methods (Chambers et al., 2023)
Comparison of ddRAD and 2b-RAD methods (Chambers et al., 2023)
Chambers et al. compared the applications of ddRAD and 2b-RAD in phylogenomics. Their study selected samples from two frog clades, namely Ranidae and Dendrobatidae, with each clade comprising five species and two individuals per species for sequencing. Both methods were able to estimate well-supported phylogenetic trees at various sequencing depths. However, the datasets generated by 2b-RAD exhibited greater reproducibility and consistency across different sequencing depths. Additionally, while the library preparation cost for 2b-RAD is lower, its overall cost is comparable to ddRAD due to the need for higher sequencing depth. Nevertheless, the advantage of 2b-RAD in handling shorter fragment lengths may render it more cost-effective in certain scenarios.
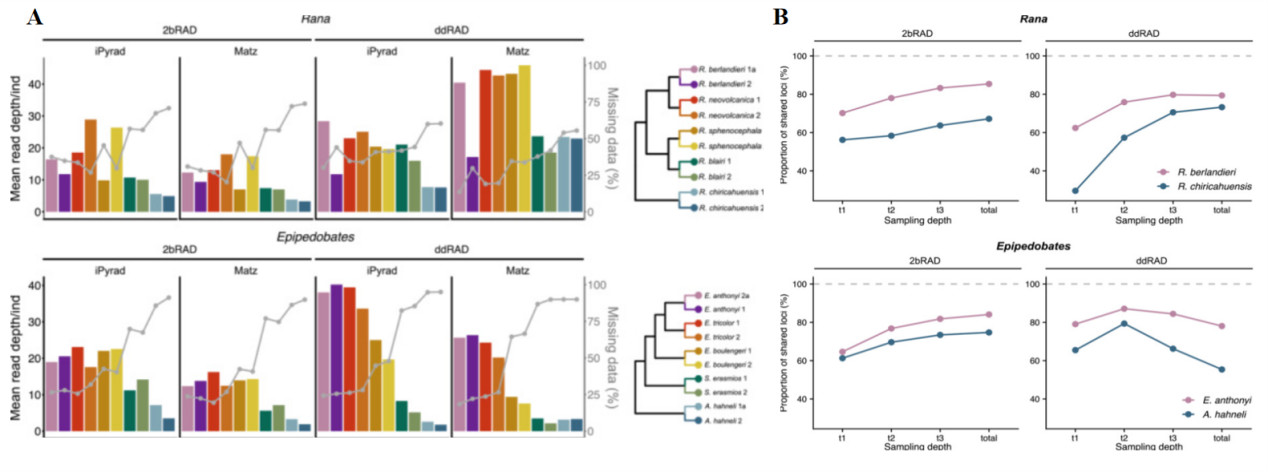 Comparison of ddRAD and 2b-RAD sequencing total SNP dataset depth and reproducibility of sequencing(Chambers et al., 2023)
Comparison of ddRAD and 2b-RAD sequencing total SNP dataset depth and reproducibility of sequencing(Chambers et al., 2023)
2b-RAD in Plant Breeding and Agriculture
In the pursuit of advancing plant breeding and agricultural development, researchers have tirelessly sought efficient and precise genetic analysis techniques. Among the array of technologies available, 2b-RAD (a simplified genome sequencing technique) stands out for its unique advantages, such as relative ease of operation, lower costs, and the ability to provide rich genetic information. It offers new opportunities and breakthroughs for plant breeding and agricultural research. Not only can it delve deeply into the genetic diversity of plants, aiding in the precise identification of desirable traits, but it also plays a crucial role in crop improvement and variety identification. Below, we will explore the practical applications of 2b-RAD in plant breeding and agriculture through specific case studies.
Zhang et al. focused on Megalobrama amblycephala and Ancherythroculter nigrocauda as their research subjects, utilizing 2b-RAD-seq technology to screen for sex-specific markers. In their study, they conducted artificially induced gynogenetic experiments on female individuals of hybrid fish, finding that all offspring were female. Subsequently, employing 2b-RAD-seq technology, they identified nine male-specific tags for the first time and successfully screened and validated two male-specific tags in hybrid and paternal A. nigrocauda populations through PCR amplification. One of the male-specific molecular markers, ref57660-1 (326 bp), was further validated through RNA-seq and quantitative analysis of gene expression levels. Combining the results that all individuals in the gynogenetic population were female with the identified male-specific markers, the study revealed that the hybrid species is homogametic female (XX) with an XX/XY sex determination mechanism. These sex-specific markers hold significant value for enhancing the efficiency of all-male/all-female breeding in this hybrid species as well as in the paternal A. nigrocauda.
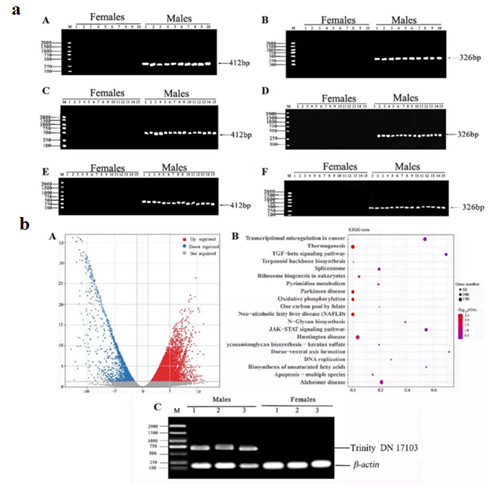 Application of 2b-RAD in Megalobrama amblycephala and Ancherythroculter nigrocauda (Zhang et al., 2022)
Application of 2b-RAD in Megalobrama amblycephala and Ancherythroculter nigrocauda (Zhang et al., 2022)
Guo et al. conducted genotyping on an F2 rice population using an improved 2b-RAD method (I2b-RAD). The study designed 12 adapters with different barcodes for adapter 1 and one adapter for adapter 2, establishing a library construction method that included digestion, ligation, pooling, PCR, and size selection. To reduce costs, non-phosphorylated adapters and indexed PCR primers were used. Through genotyping the F2 rice population, an average of 2,000,332 high-quality reads per sample were obtained, with a relatively even distribution. A total of 3,598 markers were identified, containing 3,804 SNPs, with a missing rate of 18.9%. A genetic linkage map comprising 1,385 markers was constructed, with 92% of the markers in the same order on the genetic map as on the chromosomes. Additionally, bioinformatics simulation analysis indicated that BsaX has the most extensive recognition sites across 20 species, suggesting strong potential for 2b-RAD markers in constructing high-density genetic maps. This study established an improved 2b-RAD genotyping method, I2b-RAD, which is simple, rapid, cost-effective, and suitable for multiplexed sequencing library construction. By selecting different enzymes and adapters, this method can be adjusted to suit various applications, including chromosome assembly, QTL fine-mapping, and natural population analysis.
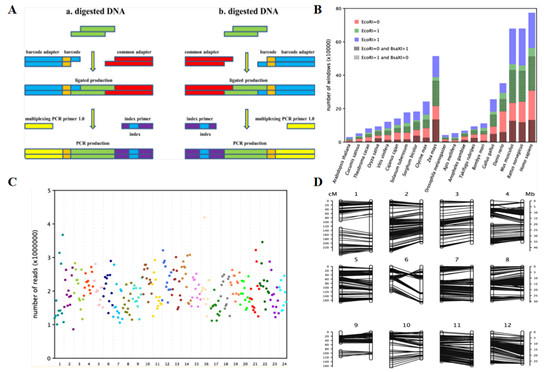 Application of 2b-RAD in rice (Guo et al., 2014)
Application of 2b-RAD in rice (Guo et al., 2014)
Ruocco et al. investigated the genetic differentiation and diversity levels of three populations of Cymodocea nodosa in the Atlantic and Mediterranean transition zones (including Gran Canaria, Faro in Portugal, and the Ebro Delta in Spain) using 2b-RAD genotyping technology. The study found significant genetic differentiation among these three populations, with varying levels of clonality and genetic diversity. The Mediterranean population of Cymodocea nodosa exhibited the highest genotype richness, while the Portuguese population had the highest clonal values, and the Gran Canaria population showed the lowest genetic diversity. Through analysis using two different methods (BayeScan and pcadapt), the researchers identified nine significant outlier single nucleotide polymorphisms (SNPs), three of which were associated with specific protein-coding genes. Among these outlier SNPs, two encoded transcription factors, and one encoded an enzyme related to flowering time regulation (lysine-specific histone demethylase 1 homolog 2), suggesting that these genes may be involved in the adaptation of seagrasses to different latitudinal environments. Furthermore, the study found that "transcriptional regulation" and "signaling" were the most abundant biological processes in the outlier SNP dataset, further emphasizing the importance of signal integration and gene regulatory networks in the adaptation of seagrass populations to environmental gradients.
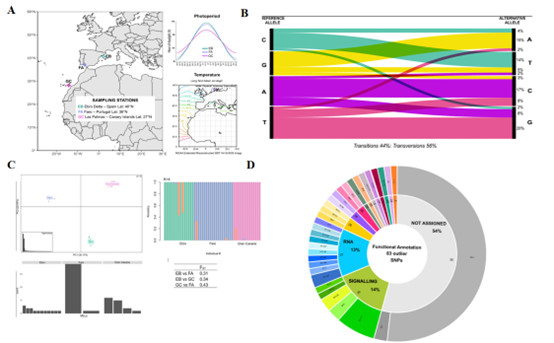 Application of 2b-RAD in Cymodocea nodosa (Ruocco et al., 2022)
Application of 2b-RAD in Cymodocea nodosa (Ruocco et al., 2022)
2b-RAD Challenges and Solutions
Technical limitations
In polyploid plants, allele dropout is a common technical challenge. Due to the complexity of polyploid plant genomes, alleles may not be accurately detected during the sequencing process. Enzyme optimization can alleviate this issue to some extent. Researchers can select enzymes more suitable for polyploid plants for 2b-RAD sequencing, improving digestion efficiency and accuracy, and reducing the occurrence of allele dropout. For example, by screening different types of restriction enzymes, researchers can identify enzymes that produce more uniform fragments in polyploid plant genomes, thereby enhancing sequencing quality.
Compatibility with low-quality DNA is another challenge faced by 2b-RAD technology. In some cases, such as with ancient seed samples, DNA may degrade and be of low quality. To address this, researchers have developed 2b-RAD protocols suitable for degraded samples. These protocols include optimizing DNA extraction methods, adjusting digestion conditions, and sequencing parameters to improve the success rate of sequencing low-quality DNA. Through these methods, even degraded DNA extracted from ancient seeds can be sequenced using 2b-RAD, offering possibilities for the study of ancient plants.
Data analysis innovations
Reference-free genome methods represent an important innovative direction in 2b-RAD data analysis. For many wild plant populations, reference genome sequences are lacking, making traditional reference-based analysis methods difficult to apply. SNP detection methods for wild plant populations based on K-mers can detect SNPs without a reference genome. A K-mer refers to a continuous subsequence of length k in a DNA sequence. By analyzing the frequency and distribution of K-mers in different samples, SNP sites can be accurately detected. This method provides an effective means for genetic research in wild plant populations, helping to reveal their genetic diversity and evolutionary history.
Integration with phenomics is another significant trend in 2b-RAD data analysis. Phenomics is the study of an organism's phenotypic traits and their interactions with the environment. Associating 2b-RAD markers with trait data obtained by drones enables comprehensive analysis of genotypes and phenotypes. Drones can efficiently capture phenotypic data of plants, such as plant height and leaf area. By combining 2b-RAD markers with these phenotypic data, researchers can gain deeper insights into the relationship between a plant's genetic mechanisms and phenotypic traits, providing more comprehensive information for plant breeding and variety improvement.
Future Directions of 2b-RAD in Plants
Hybrid approaches
Combining 2b-RAD with nanopore sequencing represents a promising hybrid method. Nanopore sequencing technology offers advantages such as real-time sequencing and long read lengths, but it has limitations in terms of accuracy and throughput. On the other hand, 2b-RAD technology is characterized by its high throughput and accuracy. By integrating these two technologies, field-deployable genotyping can be achieved. For instance, in remote areas or wild environments, researchers can utilize nanopore sequencing for rapid preliminary sequencing and then employ 2b-RAD technology for further accurate genotyping. This approach provides a more flexible and efficient means for plant genomics research, facilitating plant studies in diverse environments.
Microbiome interactions
The interactions between plants and microorganisms significantly impact plant growth, development, and the ecological environment. The 2b-RAD-M (2b-RAD for Microbiome) technology can be used to analyze plant-microbe associations in sustainable agriculture. This technology enables the sequencing and analysis of the genomes of plant-associated microorganisms, revealing the mechanisms of interaction between plants and microorganisms. For example, by studying the genomic information of plant rhizosphere microorganisms, we can understand how these microorganisms promote plant growth and enhance plant resilience to stress. This knowledge aids in the development of sustainable agricultural strategies based on plant-microbe interactions, improving agricultural production efficiency and ecological environmental quality.
In summary, 2b-RAD sequencing technology holds significant application value and development prospects in plant genomics. By continuously optimizing technical protocols, addressing challenges, and exploring future directions, 2b-RAD technology will bring more breakthroughs and innovations to plant research and breeding practices, driving the advancement of plant science.
References
- Wang S, Meyer E, et al. "2b-RAD: A Simple and Flexible Method for Genome-Wide Genotyping." Nat Methods. 2012 9(8):808-10 https://doi.org/10.1038/nmeth.2023
- Chambers EA, Tarvin RD, et al. "2b or Not 2b? 2bRAD Is an Effective Alternative to ddRAD for Phylogenomics." Ecol Evol. 2023 13(3):e9842 https://doi.org/10.1002/ece3.9842
- Si-Qi Zhang, Xue-Mei Xiong, et al. "Screening and Characterization of Sex-Specific Markers for the Hybrid (Megalobrama amblycephala ♀ × Ancherythroculter nigrocauda ♂) Based on 2b-RAD and Transcriptome Sequencing." Aquaculture. 2022 548(2):737704https://doi.org/10.1016/j.aquaculture.2021.737704
- Guo Y, Yuan H, et al. "An Improved 2b-RAD Approach (I2b-RAD) Offering Genotyping Tested by a Rice (Oryza sativa L.) F2 Population." BMC Genomics. 2014 15(1):956https://doi.org/10.1186/1471-2164-15-956
- Ruocco M, Jahnke M, et al. "2b-RAD Genotyping of the Seagrass Cymodocea nodosa Along a Latitudinal Cline Identifies Candidate Genes for Environmental Adaptation." Front Genet. 2022 13:866758https://doi.org/10.3389/fgene.2022.866758
 Send a Message
Send a MessageFor any general inquiries, please fill out the form below.