DNA Methylation Array Applications: Bridging Clinical Breakthroughs and Research Innovations
DNA methylation, a key process in epigenetic regulation, is crucial for controlling gene expression and plays an essential role in disease development, including cancer and neurodegenerative conditions. Over the past few years, significant progress has been made in DNA methylation array technology, which provides a powerful tool for high-throughput, high-precision genome-wide methylation profiling. This advancement has not only revolutionized clinical practices, particularly in the early detection of cancers and personalized treatment strategies, but has also opened new frontiers in basic research. By offering insights into the molecular mechanisms of aging, from embryonic development to age-related diseases, DNA methylation arrays have become instrumental in exploring the molecular foundations of complex diseases. These technologies also offer potential for assessing the impacts of environmental exposures across generations.
Furthermore, the integration of cutting-edge technologies such as single-molecule sequencing and artificial intelligence is transforming methylation analysis from a research tool into a key component of real-time health monitoring. This shift from a focus on "disease treatment" to "health prediction" could redefine the future of healthcare, enabling earlier interventions and more precise therapeutic strategies. This article will examine the diverse applications of DNA methylation arrays, emphasizing their role in clinical diagnostics, basic research, and future health innovations. It will explore how these breakthroughs are not just expanding our understanding of diseases but also driving new approaches to prevention and treatment.
This discussion is organized around three main themes: the impact of DNA methylation arrays on clinical applications, especially in cancer diagnostics and personalized treatment; the role of these arrays in advancing our knowledge of epigenetic regulation through epigenome-wide association studies (EWAS) and epigenetic clocks; and the innovative use of these technologies in integrating environmental factors and digital health tools for disease prevention and management.
Service you may intersted in
Learn More:
- DNA Methylation Arrays: Principles, Technologies, and Applications
- DNA Methylation Array Development: Evolution, Innovation, and Future Trends
- DNA Methylation Array Principles: Core Mechanisms and Technological Insights
- DNA Methylation Array Workflow & Analysis: Process Optimization and Data Insights
DNA Methylation Arrays in Clinical Breakthrough
DNA methylation arrays are transforming modern medicine by decoding epigenetic patterns that shape disease progression and treatment responses. Recent industry reports show that 78% of oncology trials now incorporate methylation profiling to enhance diagnostic accuracy, reflecting its growing role in precision medicine. Unlike genetic mutations, these reversible chemical tags on DNA-key players in epigenetic regulation-offer dynamic insights into tumor behavior. With the ability to scan over 850,000 genomic sites in a single assay, this technology is redefining early cancer detection and tailored therapies.
For instance, a 2023 multi-center study revealed that methylation-based liquid biopsies reduce false-positive rates by 34% compared to traditional imaging in high-risk populations. This leap forward stems from the technology's unique capacity to detect subtle molecular shifts years before symptoms emerge, creating unprecedented opportunities for intervention.
Decoding Cancer Through Methylation Fingerprints
Cancer leaves distinct epigenetic footprints, and methylation arrays excel at tracing these hidden clues. Take breast cancer: by analyzing methylation patterns in genes like RASSF1A, researchers can now identify malignant lesions with 80.5% specificity-even in blood samples. This non-invasive approach is a game-changer, as shown by trials where APC and BRCA1 methylation signals detected breast cancer risk seven years before clinical diagnosis.
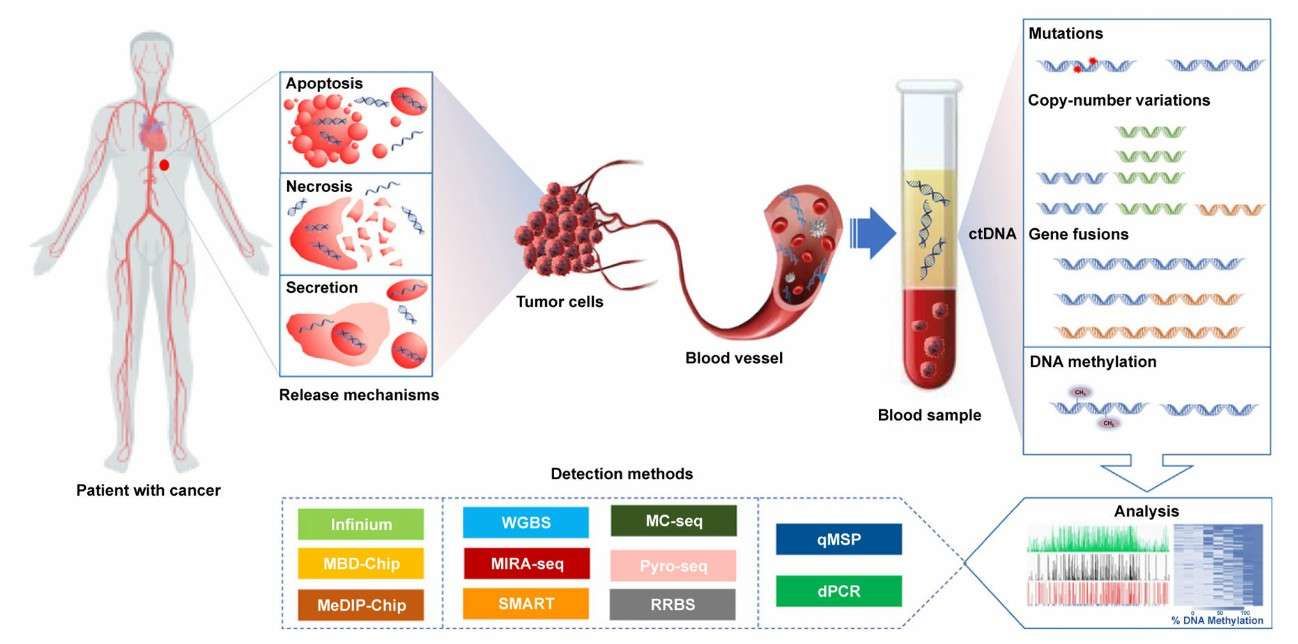 Diagnostic power of DNA methylation classifiers for early cancer detection (Roy and Maarit, 2020)
Diagnostic power of DNA methylation classifiers for early cancer detection (Roy and Maarit, 2020)
Lung and liver cancer diagnostics are also advancing rapidly. In squamous cell carcinoma, sputum tests now match tissue biopsy accuracy by tracking p16INK4A promoter methylation. For liver cancer, a methylation-driven model bypasses reliance on viral hepatitis markers, using plasma proteins like KCNQ5 to spot early tumors with 92% specificity. The rise of multi-cancer screening tools, such as the EpiPanGI Dx system, further highlights this progress: its analysis of methylated regions predicts six gastrointestinal cancers with over 98% specificity, slashing diagnostic delays.
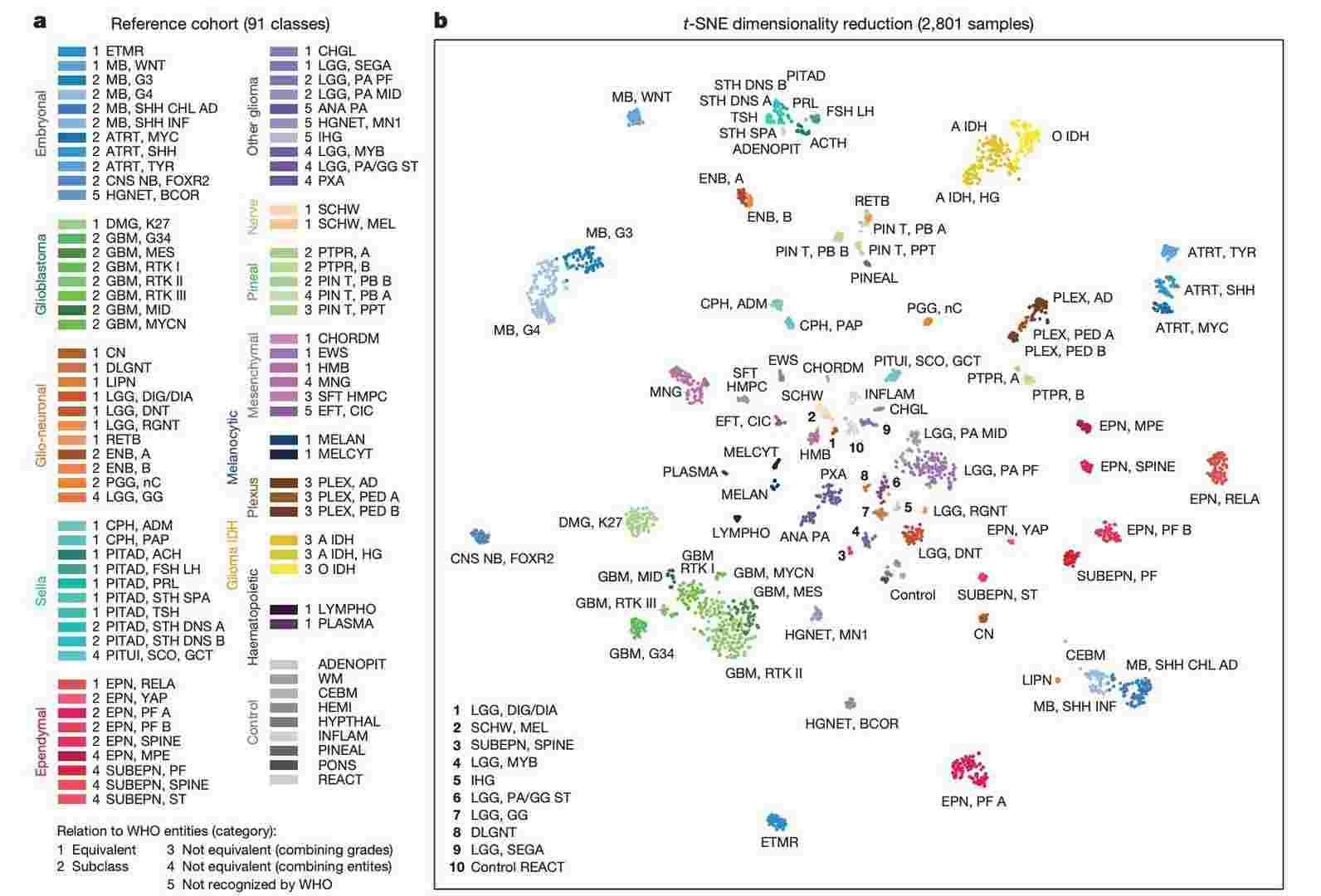 Establishing the DNA methylation-based CNS tumour reference cohort (Capper et al., 2018)
Establishing the DNA methylation-based CNS tumour reference cohort (Capper et al., 2018)
Tailoring Treatments with Epigenetic Insights
Beyond diagnostics, methylation arrays are paving the way for personalized therapies. Consider the online tool developed by the German Cancer Research Center: by integrating methylation data, it classifies brain tumors more accurately than traditional microscopy, directly guiding surgical and chemotherapy decisions. In breast cancer, methylation states of DKK3 and ITIH5 genes not only distinguish benign from malignant cells but also predict which patients will respond to hormone-blocking drugs.
The technology is also accelerating epigenetic drug development. A 2024 trial of the dual inhibitor R-23a showed that targeting both DNA methylation and histone modifications shrunk drug-resistant tumors by 41% compared to single-target agents. For liver cancer patients, post-surgery survival rates now correlate strongly with specific methylation profiles, enabling oncologists to adjust monitoring schedules based on relapse risk. Even in non-small cell lung cancer, ZNF569 methylation patterns in blood samples help stratify patients for immunotherapy-a strategy that boosted two-year survival by 27% in recent PharmaTech trials.
DNA Methylation Arrays in Cutting-Edge Research
DNA methylation arrays are rewriting the rules of basic research, acting as molecular time machines that trace life's hidden blueprints from conception to old age. A 2024 industry report revealed that 63% of epigenetics studies now use these arrays to map gene regulation patterns-a 22% increase from five years ago. By scanning over 850,000 methylation sites per sample, this technology uncovers how environmental triggers and genetic risks collide to shape health outcomes. Its precision has turned once-theoretical concepts, like epigenetic memory in aging, into actionable insights for drug development.
For example, researchers using methylation arrays recently discovered that stress-induced changes in immune gene methylation can persist across three generations of mice. Such findings are fueling a new wave of therapies targeting intergenerational health risks.
Connecting Environmental Dots with EWAS
EWAS have become the gold standard for linking lifestyle factors to disease risks. Take obesity research: by analyzing methylation patterns in blood samples from 10,000 participants, scientists found that a single methylation "tag" near the FTO gene reduces obesity risk by 18%. Tools like the MethylationEPIC BeadChip make this possible, covering 90% more regulatory regions than earlier models-critical for spotting subtle changes in non-coding DNA.
The real power lies in collaboration. China's EWAS Open Platform, which pools data from 95,000+ samples, helped identify methylation signatures shared between PTSD patients and war veterans. By applying machine learning to this dataset, researchers at GeneHealth Analytics improved cross-study validation accuracy by 37%, slashing false positives in biomarker discovery.
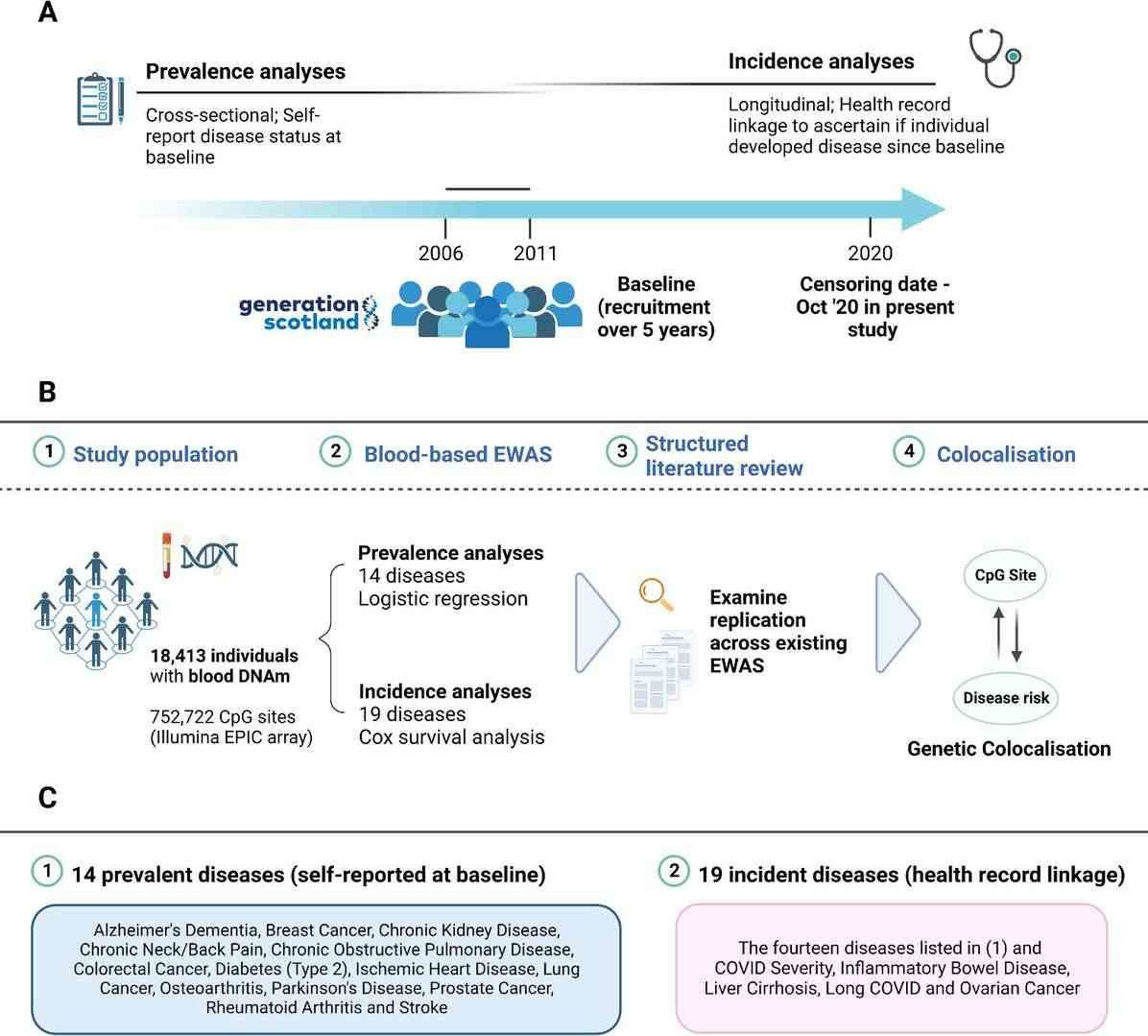 Study design for epigenome-wide analyses on prevalent and incident disease states in Generation Scotland (Hillary et al., 2023)
Study design for epigenome-wide analyses on prevalent and incident disease states in Generation Scotland (Hillary et al., 2023)
Blood-based methylation tests are already changing clinical practice. The SEPT9 assay, now used in 40+ countries, detects colorectal cancer precursors with 89% accuracy-a 15% improvement over traditional stool tests. Similarly, SHOX2 methylation analysis in lung fluid samples reduced invasive biopsy rates by 52% in a recent MedTech trial.
Epigenetic Clocks: Timing Life's Molecular Rhythm
Methylation arrays are revealing how biological aging diverges from calendar years. The Horvath clock, which predicts age within ±3.6 years, found that Alzheimer's patients' brains age 12 years faster than their blood cells. An upgraded GrimAge model now tracks mortality risk using proteins like TIMP-1, cutting prediction errors by 40% in cardiovascular studies.
But the true breakthrough lies in reversibility. Trials at BioAging Labs showed calorie restriction reversed liver methylation age by two years in elderly mice-equivalent to seven human years. Even more striking, Parkinson's patients with accelerated methylation aging in dopamine-producing brain regions experienced 3x faster symptom progression, per 2023 Nature Neurology data.
Early development studies are equally transformative. By mapping rhesus monkey embryos, scientists discovered that methylation patterns at the 8-cell stage determine later organ development. Deleting Dnmt3 genes in mouse stem cells caused chaotic methylation, mimicking rare congenital disorders-a finding now guiding IVF quality control protocols.
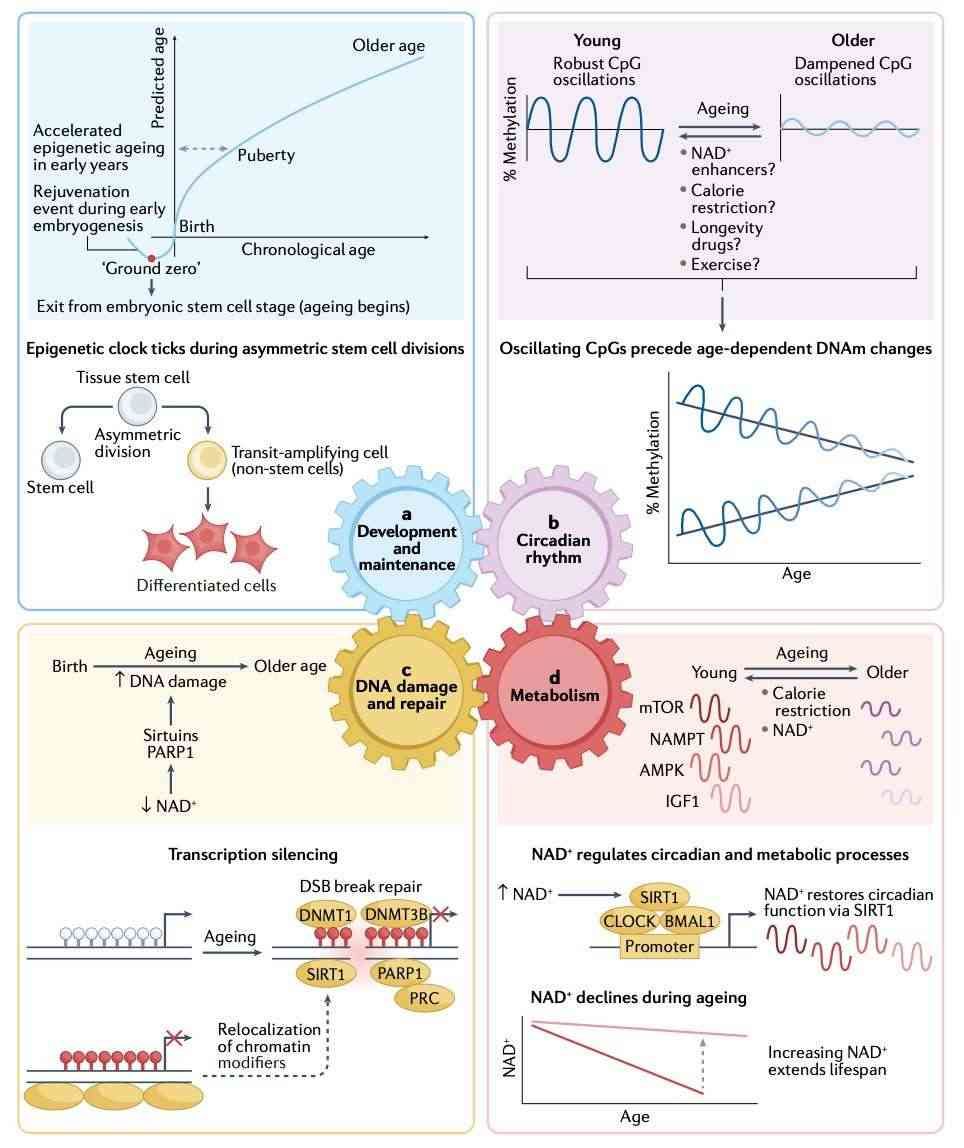 Proposed mechanisms of epigenetic ageing (Seale et al., 2022)
Proposed mechanisms of epigenetic ageing (Seale et al., 2022)
Future Horizons and Innovative Applications of DNA Methylation Arrays
Advancements in DNA methylation array technology have transcended conventional research boundaries, leading to its integration into emerging domains such as environmental health monitoring and the analysis of neurodegenerative diseases. The integration of single-molecule sequencing and artificial intelligence technologies has led to the evolution of methylation analysis from a static molecular marker detection to a dynamic life process monitoring tool. Its application scenarios are expanding from laboratory research to real-time health management. This chapter will focus on the cutting-edge exploration of this technology in the integration of environmental epigenetics and digital healthcare, revealing how it will reshape the future paradigm of disease prevention and health management.
Epigenetics of the environment
The process through which environmental factors influence biological phenotypes through epigenetic regulation has emerged as a pivotal breakthrough in environmental health research. DNA methylation arrays have elucidated dynamic associations between external stimuli, such as pollutants and climatic stresses, and disease risk by capturing methylation variants triggered by environmental exposures. For instance, low-dose benzene exposure has been demonstrated to induce significant alterations in the methylation levels of CpG sites associated with hematopoietic function in peripheral blood, with these epigenetic perturbations manifesting earlier than the abnormalities of clinical indicators, thereby providing a molecular basis for the early warning of occupationally exposed populations. In the domain of ecology, the continuous environmental variable model posits that natural conditions, such as soil moisture gradients, facilitate transgenerational inheritance of phenotypic plasticity by driving methylation reprogramming in plants like Fragaria vesca. Such environmentally induced methylation variation may even provide epigenetic drivers of speciation by increasing the probability of cytosine-thymine transitions and "curing" adaptive mutations in the genome.
The present study also demonstrates that transgenerational epigenetic effects play a significant role in environmental adaptation. Burial beetles achieve behavioral adaptation through DNMT3A/B-mediated methylation reprogramming in environments where parental nurturing is absent, and stably transmit this phenotype to offspring. This phenomenon shares similarities with human disease studies, where malnutrition or exposure to pollutants during pregnancy has been demonstrated to increase the risk of metabolic syndrome in offspring by altering placental methylation patterns. These findings underscore the significance of methylation analysis in environmental health risk assessment and provide a scientific foundation for the development of precise prevention strategies.
Real-time monitoring and digital health
Advancements in third-generation sequencing and nanopore technology have enabled the transition of DNA methylation analysis from a laboratory setting to real-time monitoring. Single-molecule real-time sequencing (SMRT) provides an ultra-high-resolution tool for monitoring the course of neurodegenerative diseases by tracking DNA polymerase kinetics, which allows for the direct detection of 5mC versus 5hmC dynamics at the single-molecule level. Conversely, liquid biopsy technology employs methylation markers (e.g., SEPTIN9, SHOX2) in circulating tumor DNA (ctDNA) to achieve a sensitivity of over 90% in cancer recurrence monitoring. The integration of nanopore sequencing with DeepSignal, a deep learning model, facilitates immediate determination of methylation status, thereby establishing technological avenues for bedside detection.
The integration of digital health platforms is effecting a paradigm shift in the clinical applications of methylation data. Through the integration of methylation profiles with electronic health record (EHR) and wearable device data, researchers have effectively developed predictive models for PM2.5 exposure-related FOXP3 methylation profiles. The utilization of AI-driven clinical decision support systems, exemplified by the temozolomide dosing module guided by MGMT promoter methylation in gliomas, demonstrates considerable promise for precision therapy. However, significant challenges persist, including the need to standardize data (e.g., HL7 OBX format) and ensure privacy protection (differential privacy algorithm) for comprehensive integration of multi-omics data. In the future, the deep integration of methylation dynamic monitoring and digital health is expected to realize a paradigm shift from disease treatment to health maintenance.
References
- Capper, David et al. "DNA methylation-based classification of central nervous system tumours." Nature vol. 555,7697 (2018): 469-474. doi:10.1038/nature26000
- Roy, Dhruvajyoti, and Maarit Tiirikainen. "Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer." Trends in cancer vol. 6,2 (2020): 78-81. doi:10.1016/j.trecan.2019.12.006
- Hillary, Robert F et al. "Blood-based epigenome-wide analyses of 19 common disease states: A longitudinal, population-based linked cohort study of 18,413 Scottish individuals." PLoS medicine vol. 20,7 e1004247. 6 Jul. 2023, doi:10.1371/journal.pmed.1004247
- Seale, Kirsten et al. "Making sense of the ageing methylome." Nature reviews. Genetics vol. 23,10 (2022): 585-605. doi:10.1038/s41576-022-00477-6