What Is 5‐Hydroxymethylcytosine
DNA methylation and hydroxymethylation are important epigenetic modifications for gene expression regulation. DNA methylation typically represses gene transcription. Hydroxymethylation, on the contrary, activates gene expression or prompts DNA demethylation.
5‐Hydroxymethylcytosine (5hmC) is a DNA modification that is generated by the oxidation of 5‐methylcytosine (5mC) in a reaction catalyzed by the ten‐eleven translocation (TET) family enzymes. It tends to mark gene activation and affects a spectrum of developmental and disease‐related biological processes.
What Is 5hmC-Seal Seq
5hmC selective chemical labeling sequencing (5hmC-Seal seq) uses the principles of click chemistry and biotin-labeled capture(Nat Biotechnol. 2011;29(1):68-72) to reduce DNA damage based on sulfite sequencing, reduce the requirement for DNA input, and meet the needs of the genome-wide hydroxymethylation detection for different sample types(cells, tissues, whole blood, plasma cell-free DNA), and is a cost-effective detection solution for DNA hydroxymethylation.
Application of 5hmC-Seal Seq
1) Research on the methylation mechanism of development and disease;
2) Molecular classification and diagnosis and prognosis of complex diseases and tumors;
3) Clinical applications such as cfDNA hydroxymethylation detection and liquid biopsy.
5hmC-Seal Seq Workflow:
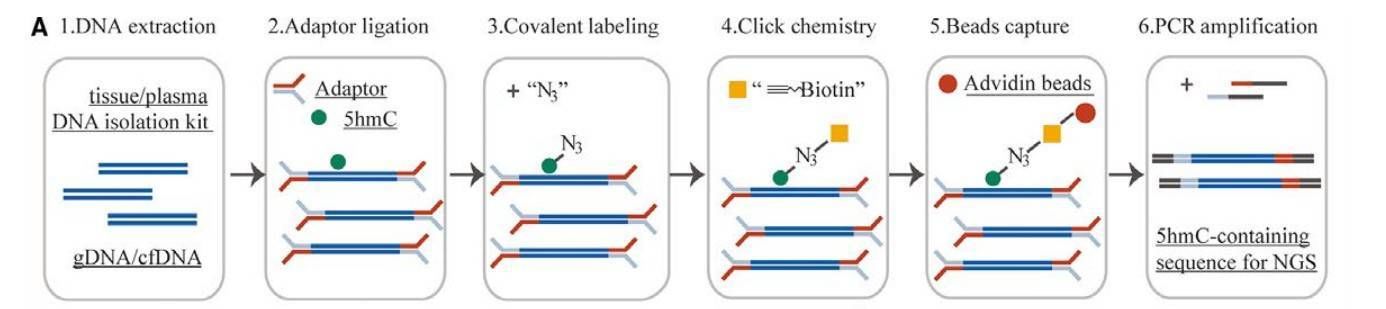
Service Specifications
Sample Requirements
|
|
 |
Sequencing Strategy
|
 |
Bioinformatics Analysis We provide customized bioinformatics analysis including:
|
Advantages of Our 5hmC-Seal Seq Service
A complete cfDNA hydroxymethylation detection solution from sample collection, cfDNA extraction to library construction and sequencing.
Core technology for 5hmC Seal library construction as low as 5ng plasma cfDNA.
Automated sample processing, library building and analysis processes to ensure an efficient cycle.
Sequencing and analysis of various tumor tissues, cfDNA and mammalian hydroxymethylation have been completed.
1: Detection of 5-hydroxymethylcytosine based on plasma cell-free DNA for early diagnosis of liver cancer
Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019 Jul 29.
Study protocol: In this paper, researchers applied 5hmc-seal technology to detect genome-wide 5hmC in 2554 Chinese cfDNA samples, including 1204 patients with liver cancer and 392 patients with chronic hepatitis B virus infection or cirrhosis and 958 healthy individuals.
RESULTS: A diagnostic model for early stage hepatocellular carcinoma was established by case-control analysis. Based on the Barcelona Clinical Liver Cancer Staging System, a 32-gene diagnostic model was developed that accurately distinguished early-stage liver cancer (stage 0/a) from non-hepatocellular carcinoma, and also well differentiated early-stage liver cancer from high-risk patients with a history of CHB or IC (AUC=84.6%, 95% CI 85.8% to 91.1%); furthermore, the 5hmC diagnostic model appeared to be associated with potential confounders, such as history of smoking/alcohol consumption. Therefore, this method has potential for clinical application in the early detection of liver cancer.
2: Hydroxymethylation in pancreatic cancer alters and regulates cancer gene pathways
Altered hydroxymethylation is seen at regulatory regions in pancreatic cancer and regulates oncogenic pathways. Cell Research. 2017;27(11):1830-1842.
Study Protocol: 5-Hydroxymethylcytosine (5-hmC) is an epigenetic modification that has not been studied in pancreatic cancer. Whole-genome sequencing analysis of 5hmC using 5hmC-seal technology in a panel of low-passage pancreatic cancer cell lines, primary patient-derived xenograft cells, and pancreatic control tissues.
Research result:
1. Differentially hydroxymethylated regions preferentially affect known genomic regulatory regions.
2. In addition, single-base resolution analysis of cytosine methylation and hydroxymethylation by oxidative bisulfite sequencing, chromatin accessibility by ATAC-seq and gene expression by RNA-seq. Studies have found that 5hmC is specifically enriched in open chromatin regions and is associated with pancreatic tumor-related oncogenic pathways, such as MYC, KRAS, VEGFA, and BRD4;
3. In most tumor samples, BRD4 was overexpressed in the enhancer region and acquired 5hmC modification;
4. In terms of function, the increase of 5hmC level in BRD4 promoter is related to the increase of its transcriptional expression level;
5. Blocking BRD4 inhibits pancreatic cancer growth in vivo. Taken together, 5hmC redistribution and preferential enrichment of oncogene enhancers is a novel regulatory mechanism in pancreatic cancer.
References
- Song C X, Szulwach K E, Fu Y, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology, 2011, 29(1): 68-72.
- Gao P, Lin S, Cai M, et al. 5‐Hydroxymethylcytosine profiling from genomic and cell‐free DNA for colorectal cancers patients. Journal of cellular and molecular medicine, 2019, 23(5): 3530-3537.
- Applebaum M A, Barr E K, Karpus J, et al. 5-Hydroxymethylcytosine profiles are prognostic of outcome in neuroblastoma and reveal transcriptional networks that correlate with tumor phenotype. JCO precision oncology, 2019, 3: 1-12.
- Li W, Zhang X, Lu X, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell research, 2017, 27(10): 1243-1257.
