Expertise in RNA Modification Analysis
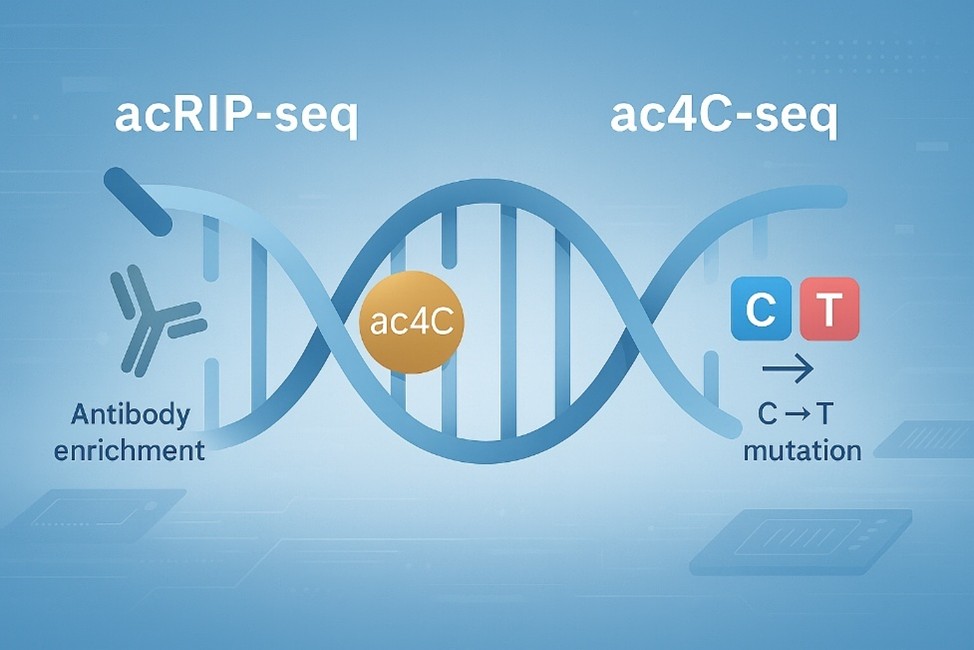
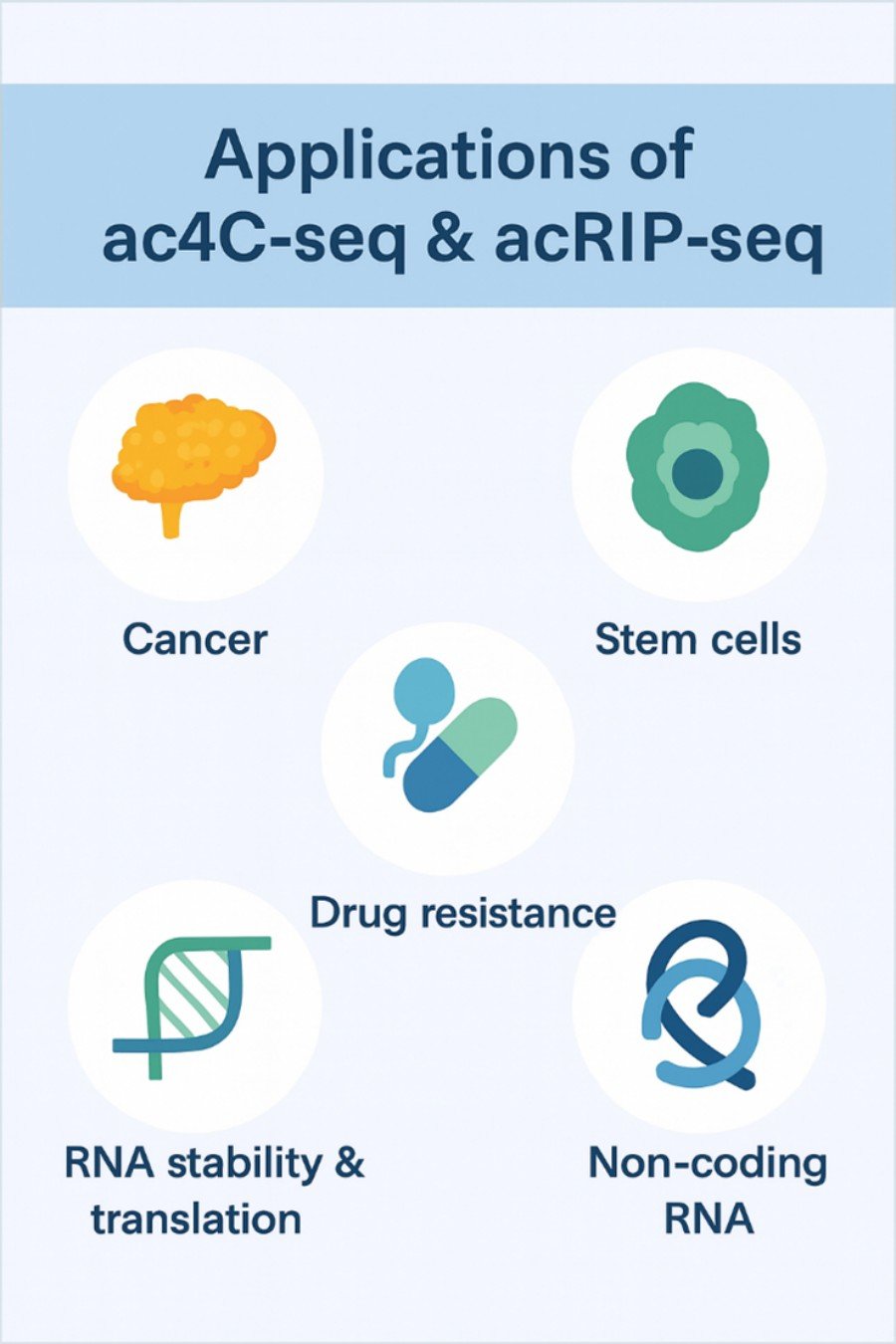
Our team lives and breathes RNA modification research—especially ac4C. We've honed our skills in both acRIP-seq and ac4C-seq through countless projects, from basic mechanistic studies to disease-focused investigations. We know the nuances of this modification inside and out, so we can deliver the precise, actionable insights you need to move your research forward.
Consistent, Reproducible Results
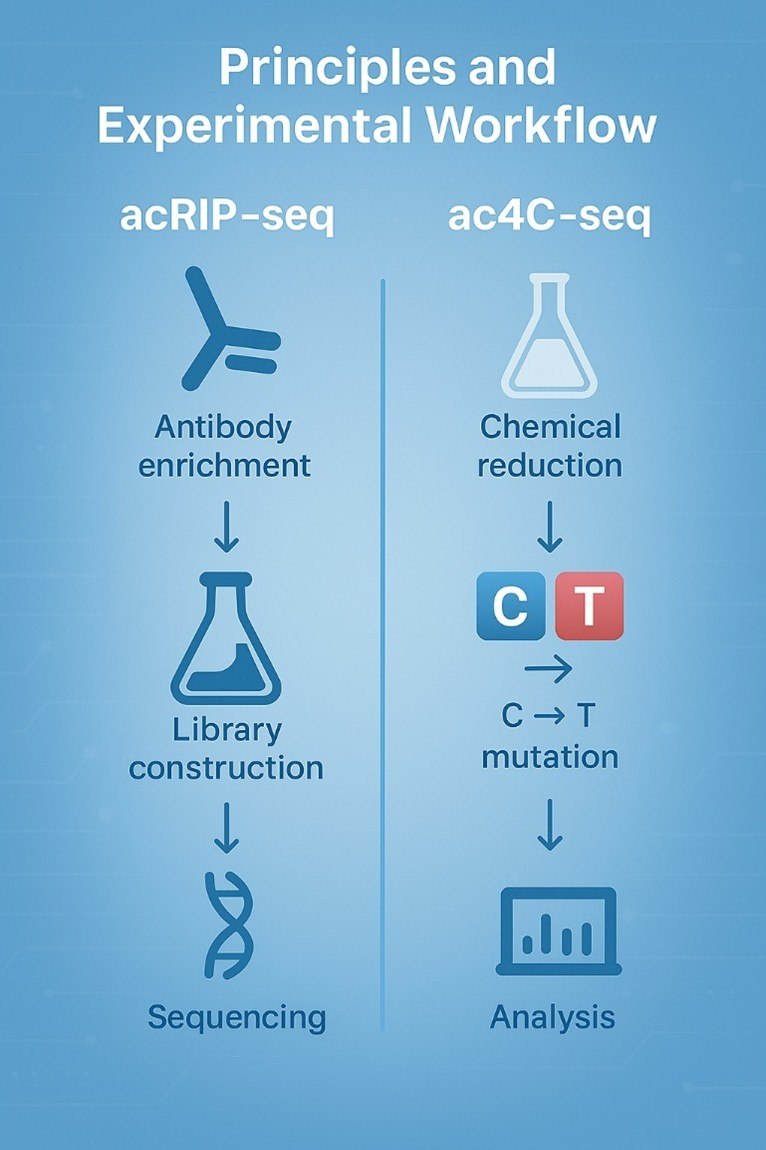
Ac4C sequencing is tricky—we get it. That's why our protocols are fine-tuned to handle its unique challenges, from antibody specificity in acRIP-seq to mutation mapping in ac4C-seq. Our meticulous approach ensures you get reliable, repeatable data every time—no surprises, no do-overs.
Custom Bioinformatics That Fits Your Project
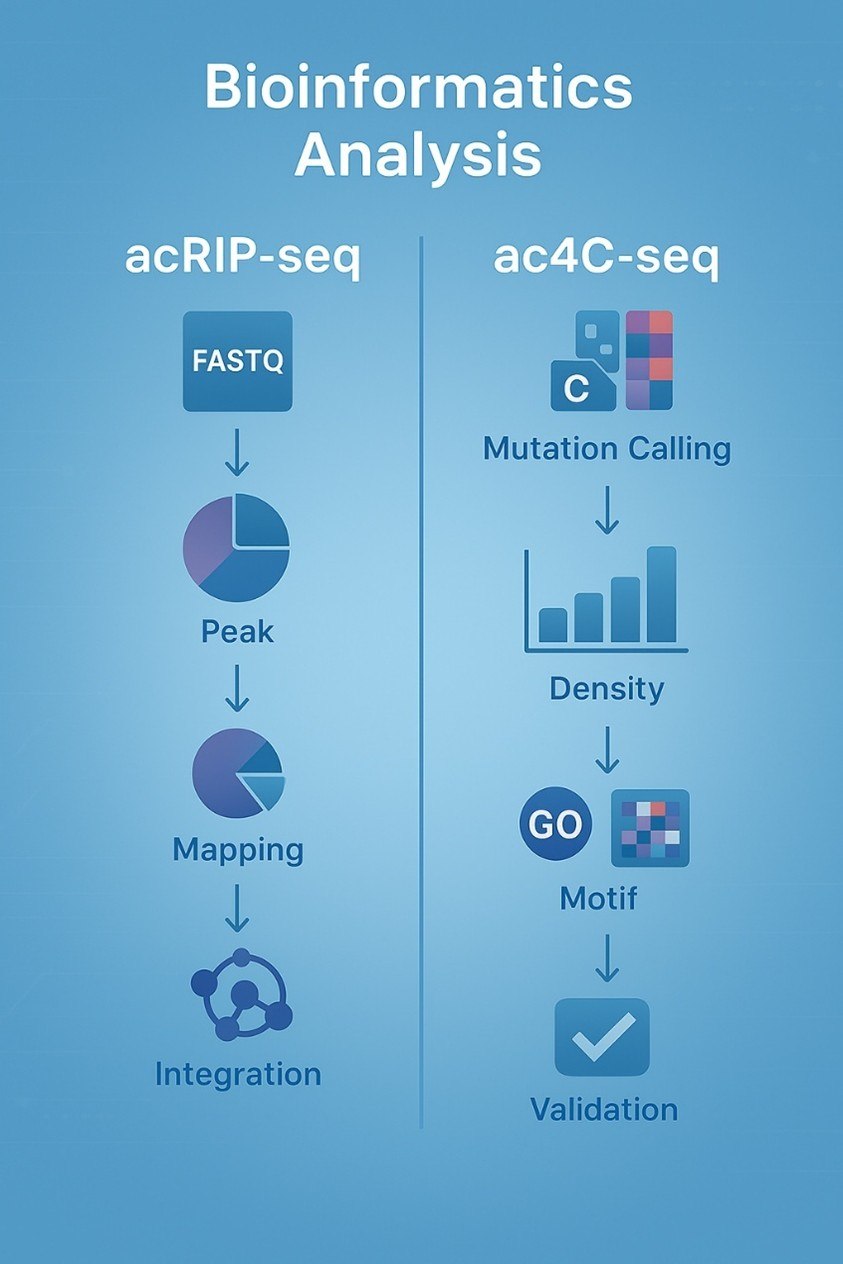
Bioinformatics isn't a box to check—it's a tool to unlock discoveries. We tailor our analyses to your goals: whether you're integrating ac4C data with RNA-seq to link modifications to expression, or using Ribo-seq to connect ac4C to translation, we'll build a pipeline that works for you. We don't just give you data—we help you make sense of it.
Clean, Publication-Ready Data
Our bioinformatics pipeline is designed to cut through noise. From peak calling to mutation validation, we prioritize quality at every step. You'll get organized, interpretable data sets—no messy files, no hidden artifacts—so you can jump straight into downstream analysis and writing.
Fast Turnaround, No Compromises
Research moves fast, and so do we. We've streamlined our workflow—from sample check-in to final results—so you get your data when you need it. We never sacrifice quality for speed: our team works efficiently to keep your project on track without cutting corners.
Scientific Rigor You Can Trust
Rigor isn't just a buzzword for us—it's how we work. We apply the same high standards that have made our work a staple in top journals. When you partner with us, you're getting results that stand up to scrutiny—because your research deserves nothing less.
We're With You Every Step of the Way
We're not just a service provider—we're your research partner. We take the time to understand your goals, whether you're generating hypotheses or validating findings. From experimental design to data interpretation, we're here to answer questions, offer guidance, and help you turn ac4C data into meaningful discoveries.