A Comprehensive Guide to RIP-Seq Technology: Workflow, Data Analysis and Experimental Design
As the core link of the gene expression regulation network, the interaction between RNA and protein plays a vital role in cell growth, differentiation, aging, and the occurrence and development of diseases. RNA Immunoprecipitation Sequencing (RIP-Seq) technology is a key tool to analyze RNA-protein interaction genomics, which skillfully combines RNA immunoprecipitation and high-throughput sequencing technology, and can systematically explore the dynamic interaction between them at the level of the complete transcriptome.
In recent years, with the iterative innovation of technology and the continuous expansion of application scenarios, RIP-Seq has made great achievements in disease mechanism analysis, new biomarker mining, and drug target screening. However, the complexity of technical principles, the meticulous requirements of experimental procedures, and the deep challenges of data analysis also mean that there are still many bottlenecks to be broken through in this field.
This paper comprehensively introduces RIP-Seq technology, including its principle, experimental process, data analysis steps, and expounds its application and value in gene expression regulation and disease research.
What is RIP-Seq
RIP-Seq technology combines RNA Immunoprecipitation (RIP) and high-throughput sequencing. RNA immune coprecipitation technology originated from protein immune coprecipitation (Co-IP). Its basic principle is to capture the RNA bound to the target protein in the cell lysate by using the specific binding of antigen and antibody, to study the interaction of RNA-protein complex in the natural state of cells.
High-throughput sequencing technology can quickly and efficiently determine a number of nucleic acid sequences. After the combination of the two, RIP-Seq can not only identify RNA interacting with specific proteins, but also perform RNA-Seq analysis on these RNAs in the whole transcriptome range, which greatly expands the depth and breadth of research.
The development of RIP-Seq technology is closely related to the demand for life science research and technical progress. In the early days, researchers used traditional RIP experiments combined with Northern Blot and RT-PCR to detect a small amount of known RNA, which was inefficient and limited in flux.
With the rapid development of high-throughput sequencing technology and the gradual integration of RIP and sequencing technology, RIP-Seq technology came into being. Nowadays, RIP-Seq has become a key technology to study RNA-protein interaction networks, which is widely used in many fields, such as gene expression regulation, disease marker screening, drug target discovery, and so on.
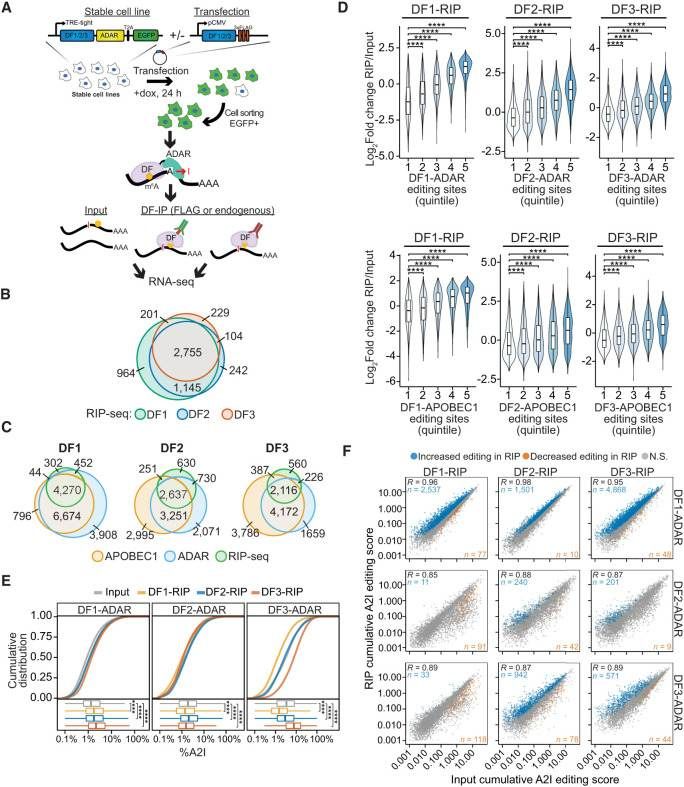 RIP-seq confirms cobinding of DF proteins to the same mRNA molecules (Flamand et al., 2022)
RIP-seq confirms cobinding of DF proteins to the same mRNA molecules (Flamand et al., 2022)
The Protocol of RIP-Seq
RIP borrows principles from protein immunoprecipitation (Co-IP) but focuses on RNA-protein complexes. The process begins with:
Cell Lysis: Gentle disruption of cells using detergents like Triton X-100, paired with protease and RNase inhibitors. Release complexes intact while preventing degradation. For sensitive cell types (e.g., primary neurons), optimized lysis buffers with reduced detergent concentrations may be necessary to avoid premature complex dissociation.
Antibody Selection: The backbone of RIP specificity. Monoclonal antibodies (mAbs) are prized for their high selectivity, minimizing non-specific binding—a critical factor in reducing background noise. Polyclonal antibodies, while more promiscuous, can capture multiple epitopes on the target protein, enhancing recovery rates for low-abundance interactors.
Immunoprecipitation: After antibody-incubation, Protein A/G beads (or magnetic beads for high-throughput workflows) capture the antibody-protein-RNA complexes. Stringent washing protocols follow, using buffers with varying salt concentrations (e.g., low-salt for weak interactors, high-salt for stringent purification) to remove unbound molecules without disrupting the complexes.
Once RNA is isolated from complexes, sequencing transforms raw molecules into biological insights:
RNA Fragmentation: Long RNA transcripts (e.g., mRNAs, lncRNAs) are sheared into 100–300 bp fragments. Chemical fragmentation (e.g., zinc-induced hydrolysis) offers random cleavage, while enzymatic methods (e.g., RNase III) produce more uniform sizes. The choice depends on downstream applications—random fragmentation is ideal for transcriptome-wide mapping, while enzymatic methods suit targeted analyses.
Library Construction: A multi-step process to prepare fragments for sequencing.
- End Repair: Corrects damaged 5' and 3' ends, adding phosphate groups for adapter ligation.
- A-Tailing: Adds a single adenine to the 3' end, facilitating adapter binding via complementary thymine overhangs.
- Adapter Ligation: Attaches sequencing adapters (index for multiplexing) to fragments. Molecular barcodes can be incorporated here to quantify PCR duplicates and enhance accuracy.
- PCR Amplification: Amplifies library molecules to detectable levels, with cycle numbers tightly controlled to avoid bias (typically 10–15 cycles for low-input samples)
Sequencing Platforms: Illumina's NovaSeq and HiSeq dominate due to their accuracy (99.9% base call accuracy) and scalability. For rare cell populations or low-abundance RNAs, higher sequencing depths (e.g., 50–100 million reads per sample) are recommended to ensure statistical power.
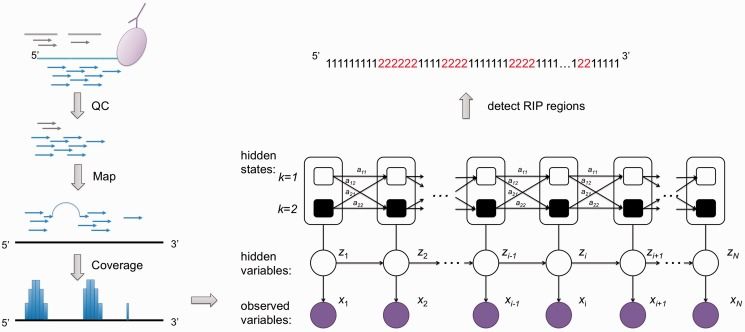 Simplified biological principles of RIP-seq (Li et al., 2013)
Simplified biological principles of RIP-seq (Li et al., 2013)
Service you may intersted in
Learn More:
How to Analyse RIP-Seq Data
RIP-Seq generates terabytes of data, requiring a robust analytical pipeline to transform raw sequences into biological meaning. Below is a step-by-step breakdown.
Quality Control
The original sequencing data often contains low-quality sequences, linker contamination, and sequencing errors, so it is necessary to control the quality of the data first.
- Raw Data Assessment: Tools like FastQC provide instant insights into data quality—look for consistent base quality scores (>Q30), balanced GC content, and minimal adapter contamination. A sudden drop in quality at read ends may indicate RNase activity during sample preparation.
- Trimming & Filtering: Trimmomatic or Cutadapt removes adapters, low-quality bases (Phred score <20), and short reads (<50 bp). Aim for a minimum of 80% clean reads post-processing; lower rates may signal issues in library preparation.
Mapping Reads to the Reference Genome
After obtaining clean data, it is necessary to compare the sequencing reads to the reference genome or transcriptome.
- Alignment Tools: Bowtie2 (for speed) and STAR (for splice-aware mapping of eukaryotic RNAs) are popular choices. For novel transcripts or alternative splicing events, aligners like HISAT2 offer superior sensitivity.
- Metrics to Monitor: Track alignment rate (≥90% for mammalian genomes), unique mapping rate (≥70%), and strand specificity (critical for lncRNA analysis). Poor alignment may stem from outdated reference genomes or RNA fragmentation artifacts.
Quantification and Normalization
Gene expression quantification of RIP-Seq data is an important step to analyze the RNA abundance bound to the target protein.
- Gene-Level Counting: featureCounts or HTSeq count reads mapping to genes, exons, or transcripts. For multi-exonic genes, HTSeq's intersection-strict mode ensures reads are assigned to the correct transcript isoform.
- FPKM (Fragments Per Kilobase of Exon per Million Mapped Reads): Adjusts for gene length and sequencing depth, ideal for comparing expression across genes within a sample.
- TPM (Transcripts Per Million): Normalizes for both gene length and library size, enabling direct comparisons between samples.
Differential Expression Analysis
Differential expression analysis aims to find out the genes with significant differences in RNA expression binding to the target protein between the experimental group and the control group. Tools like DESeq2 or edgeR identify RNAs with significantly altered binding to the target protein between conditions (e.g., treated vs. untreated cells).
- Biological Replicates: Include at least 3 replicates per group to account for biological variability; fewer replicates increase false discovery rates.
- Thresholds: Use adjusted p-values (e.g., Benjamini-Hochberg FDR <0.05) and fold changes (≥2 or ≤0.5) to filter candidates. Volcano plots (ggplot2) visualize results, highlighting high-impact RNAs.
Functional Enrichment
After screening differentially expressed RNA, it is very important to further explore its biological function.
- GO and KEGG Analysis: clusterProfiler or Enrichr map differentially bound RNAs to Gene Ontology (GO) terms (biological processes, cellular components, molecular functions) and KEGG pathways. For example, enrichment in "RNA splicing" GO terms may suggest the target protein regulates pre-mRNA processing.
- Network Construction: Cytoscape or STRINGDB can build protein-RNA interaction networks, integrating publicly available datasets (e.g., ENCODE, GTEx) to prioritize hub molecules.
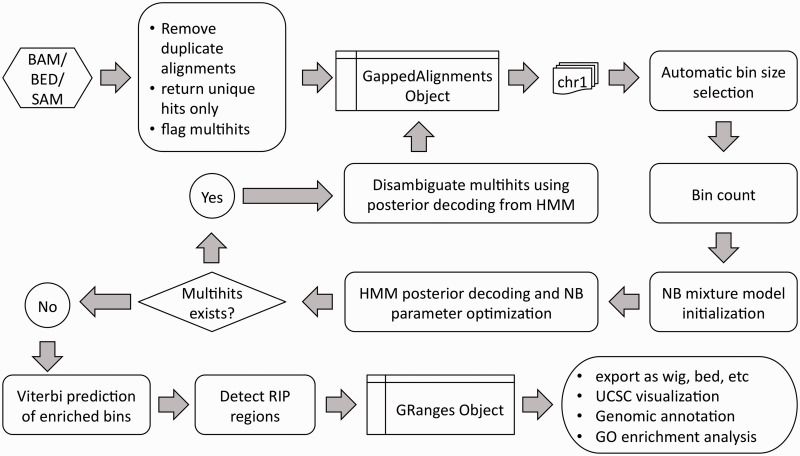 Detailed workflow of RIPSeeker (Flamand et al., 2022)
Detailed workflow of RIPSeeker (Flamand et al., 2022)
Validation and Visualization
After completing the above analysis, various visualization tools (such as ggplot2, Circos, etc.) are used to display the analysis results in the form of charts, which can present the data characteristics and laws more intuitively.
- qPCR: Quantifies binding enrichment for select RNAs using primers flanking immunoprecipitated regions. Compare Ct values between IP and Input samples to calculate fold enrichment.
- RNA Pull-Down: Biotin-labeled RNA baits capture interacting proteins, followed by Western blotting or mass spectrometry. Ideal for confirming direct interactions.
- CLIP-Seq: Crosslinking and immunoprecipitation with high-throughput sequencing offers nucleotide-resolution mapping, validating binding sites identified by RIP-Seq.
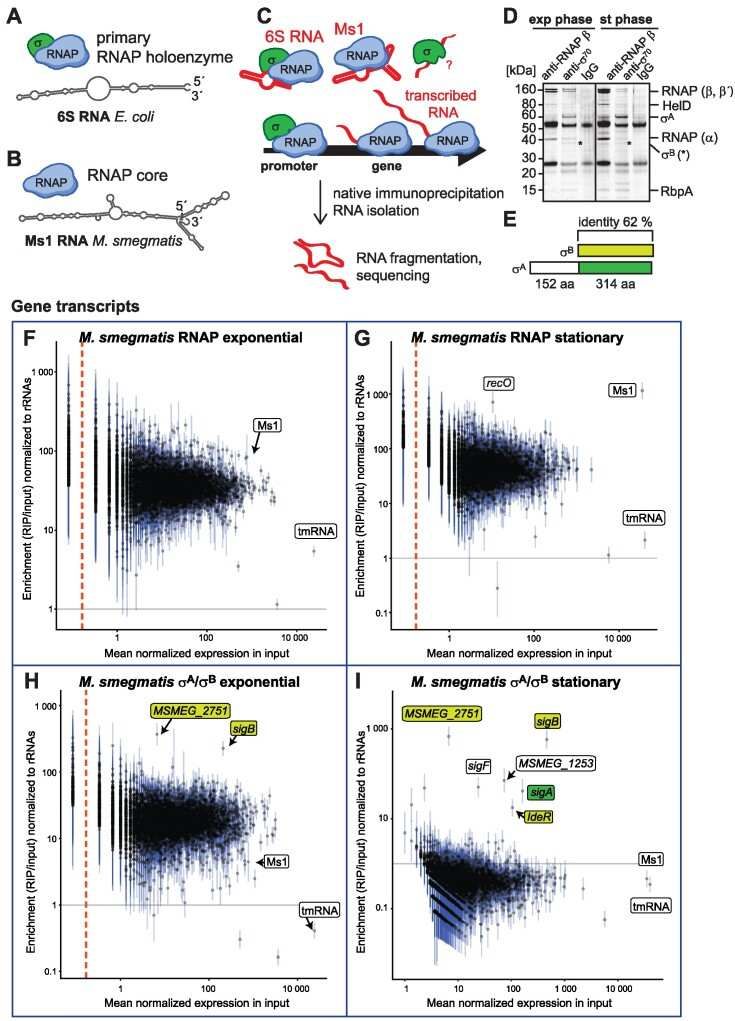 RIP-seq in Mycobacterium smegmatis (Vaňková et al., 2024)
RIP-seq in Mycobacterium smegmatis (Vaňková et al., 2024)
Service you may intersted in
RIP-Seq Comparative Experimental Design
In the RIP-Seq experiment, the key control experiment design is the core link to ensure the reliability of the results. It eliminates non-specific interference and verifies the validity of the experiment by setting a reasonable contrast, just like a "ruler" to measure the reliability of data.
IgG Negative Control
IgG control is an important negative control in the RIP-Seq experiment. IgG is a kind of immunoglobulin and does not have the ability to specifically recognize the target protein.
- Purpose: Measures non-specific binding from beads, antibodies, or reagents. Use a matched isotype control (e.g., mouse IgG for mouse-derived primary antibodies) to mimic experimental conditions without target specificity.
- Interpretation: If IgG controls show high enrichment for "background" RNAs (e.g., ribosomal RNAs, tRNAs), suspect bead contamination or overly permissive washing conditions. Pro tip: Pre-clear lysates with protein A/G beads before adding antibodies to reduce nonspecific binding.
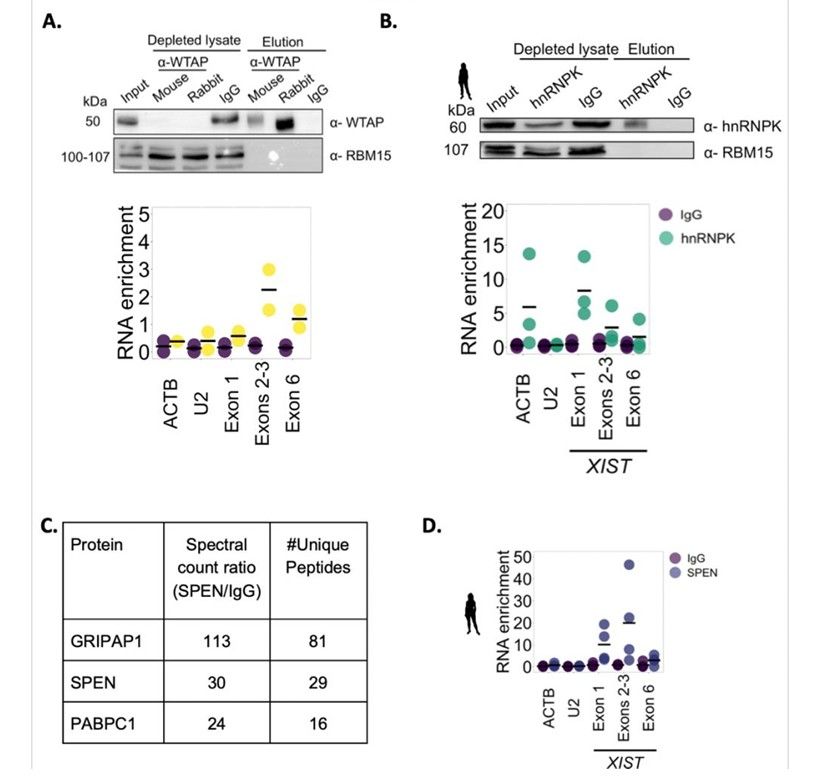 Human XIST interacts with WTAP, HNRNPK and SPEN (Ioannis et al., 2023)
Human XIST interacts with WTAP, HNRNPK and SPEN (Ioannis et al., 2023)
Input RNA Control
Input RNA control refers to RNA samples directly extracted from cell lysate before RNA immunoprecipitation. This sample represents the overall situation of all RNA in the cell and serves as a reference standard for the experiment. The library of Input RNA samples was constructed and sequenced, and compared with the sequencing results of the experimental group samples, the enrichment multiple of the target RNA in the immunoprecipitation process could be calculated. By analyzing the enrichment multiple, we can:
- Calculate enrichment fold: (IP reads / Input reads) normalizes for RNA abundance.
- Detect RNA loss during processing: If Input RNA yields low sequencing reads, investigate lysis efficiency or RNase contamination.
- Calibrate differential expression: Input data can correct for global RNA expression changes between samples.
Conclusion
To sum up, RIP-Seq technology provides a powerful technical means for studying RNA-protein interaction by skillfully combining RNA immunoprecipitation with high-throughput sequencing. During the experiment, every step from cell lysis, antibody selection, to library sequencing, as well as the design of key control experiments such as IgG control and Input RNA control, all play a decisive role in the accuracy and reliability of the experimental results. With the continuous development and improvement of technology, RIP-Seq technology will play a more important role in the field of life science research and reveal more mysteries of life phenomena for us.
References
- Flamand MN, Ke K, Tamming R, Meyer KD. "Single-molecule identification of the target RNAs of different RNA binding proteins simultaneously in cells." Genes Dev. 2022 36(17-18) :1002-1015 https://doi.org/10.1101/gad.349983.122
- Li Y, Zhao DY, Greenblatt JF, Zhang Z. "RIPSeeker: a statistical package for identifying protein-associated transcripts from RIP-seq experiments." Nucleic Acids Res. 2013 41(8): e94 https://doi.org/10.1093/nar/gkt142
- Vaňková Hausnerová V, Shoman M., et al. "RIP-seq reveals RNAs that interact with RNA polymerase and primary sigma factors in bacteria." Nucleic Acids Res. 2024 52(8): 4604-4626 https://doi.org/10.1093/nar/gkae081
- Ioannis Tsagakis, Haidee Tinning., et al. "Differences in binding preferences for XIST partners are observed in mammals with different early pregnancy morphologies." bioRxiv. 2023 https://doi.org/10.1101/2023.10.04.560814
- Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. "The RNA modification landscape in human disease." RNA. 23(12): 1754-1769 https://doi.org/10.1261/rna.063503.117




