Comparative Insights into RIP-Seq and CLIP-Seq: From Basics to Applications
In the research of gene expression regulation, the interaction between RNA and protein plays an important role. RNA immunoprecipitation sequencing (RIP-Seq) and cross-linked immunoprecipitation sequencing (CLIP-Seq), as two core technologies, provide powerful tools for analyzing RNA-protein interaction (RPI). Although both aim to capture RNA molecules bound to specific proteins, there are significant differences in technical principles, application scenarios and data characteristics. A deep understanding of these similarities and differences will help researchers choose appropriate methods according to scientific problems and accurately reveal the regulation mechanism of RPI in physiological and pathological processes.
In this paper, the differences between RIP-Seq and CLIP-Seq in technical principle, experimental process, resolution, sensitivity and application are compared and analyzed to guide researchers choose appropriate technologies according to their needs.
Brief Introduction of RIP-Seq and CLIP-Seq
RIP-Seq technology is based on the principle of antigen-antibody specific binding, aiming at capturing RNA-protein complexes naturally existing in cells. Its development stems from the improvement of traditional RNA immunoprecipitation (RIP) technology, and combined with high-throughput sequencing technology, the binding target of RBP is identified at the level of complete transcriptome.
- Firstly, the RNA-protein complex bound to a specific RBP was precipitated from the cell lysate by using specific antibodies.
- Then the precipitated RNA was purified, fragmented, reverse transcribed and high-throughput sequencing.
- The binding site and target RNA of RBP were determined by bioinformatics analysis.
RIP-Seq can well reflect the physiological state of RNA-protein interaction in cells, and is especially suitable for studying dynamic and instantaneous interactions.
The core of CLIP-Seq technology is to covalently cross-link RNA-protein by ultraviolet (UV) irradiation to form a stable complex, thus fixing the interaction of RNA-protein in cells. This technology is developed on the basis of RIP technology. By introducing ultraviolet cross-linking step, the problem that it is difficult for traditional RIP technology to accurately identify the direct binding site of RNA-protein is solved.
After a series of treatments, such as immunoprecipitation, RNA purification, reverse transcription, etc., the crosslinked complex was subjected to high-throughput sequencing, and combined with bioinformatics analysis, the binding site of RBP and RNA could be accurately located at the single nucleotide level. CLIP-Seq is suitable for studying the stable RNA-protein interaction, especially in accurately identifying RNA-protein binding sites.
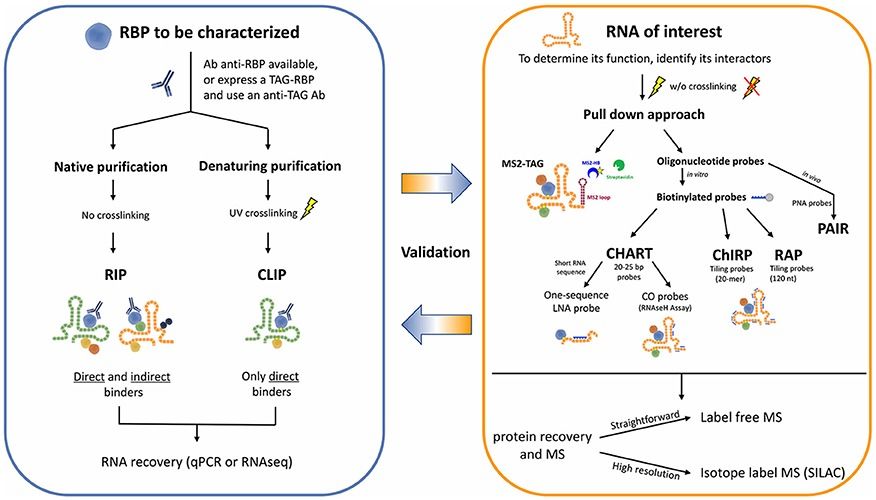 Workflow for the identification of RNA-protein interactions (Barra et al., 2017)
Workflow for the identification of RNA-protein interactions (Barra et al., 2017)
Service you may intersted in
Learn More:
Key Differences Between RIP-Seq and CLIP-Seq
RIP-Seq and CLIP-Seq are both core technologies for studying RNA-protein interaction, but there are significant differences in process, sensitivity and so on.
Experimental Process Difference
- Crosslinking step
- a)The cross-linking step is one of the key differences between RIP-Seq and CLIP-Seq. Before cell lysis, CLIP-Seq used UV irradiation to form covalent cross-linking between RNA and RBP. This cross-linking method can stabilize the RNA-protein interaction, and even in the subsequent violent operations such as cell lysis and immunoprecipitation, it can retain the combined state of the two to the greatest extent, thus ensuring that the detected RNA-protein interaction is a real direct combination in the cell. However, UV crosslinking may also have some disadvantages, such as destroying the normal structure of some conformation-sensitive RBPs and affecting their binding specificity with RNA.
- b)In contrast, RIP-Seq directly utilizes the naturally occurring RNA-protein interaction in cells without UV cross-linking. Cells were lysed under relatively mild conditions, and then RNA-protein complexes were captured by specific antibodies. This method can well simulate the physiological environment of RNA-protein interaction in cells, and has advantages in studying dynamic and instantaneous interactions. However, due to the lack of cross-linking immobilization, some weakly interacting or instantaneously bound RNA-protein complexes may be dissociated during the experiment, which affects the sensitivity of detection.
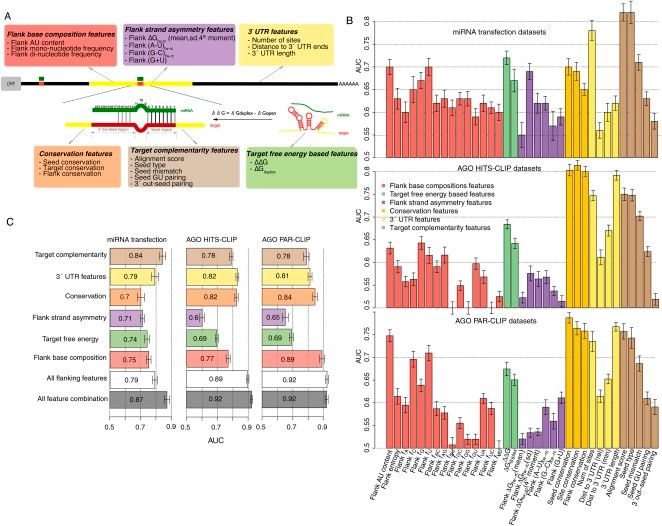 Predictive power of both individual miRNA-target features and feature combinations (Wen et al., 2011)
Predictive power of both individual miRNA-target features and feature combinations (Wen et al., 2011)
- Antibody demand
- a)In terms of antibody requirements, RIP-Seq has stricter requirements on antibody specificity. Because RIP-Seq relies on the specific binding of antibody and RBP to capture RNA-protein complex, if the specificity of antibody is insufficient, it will easily lead to non-specific binding, which will introduce a lot of background signals and affect the accuracy of experimental results. Therefore, in RIP-Seq experiments, it is usually necessary to use highly specific antibodies that have been strictly verified. In addition, in order to improve the reliability of the experiment, a variety of antibodies can be used for parallel experiments to verify the experimental results.
- b)Although CLIP-Seq also needs to use specific antibodies, due to the existence of UV cross-linking step, RNA covalently cross-linked with RBP will be precipitated together even if there is a certain degree of non-specific binding of antibodies, and the non-specific signals can be filtered and eliminated by bioinformatics methods in the subsequent analysis. However, in order to obtain more accurate experimental results, we should also try to choose highly specific antibodies in the CLIP-Seq experiment.
Comparison of Resolution and Sensitivity
The biggest advantage of CLIP-Seq is that it can identify RNA-protein binding sites with single nucleotide resolution. Because the covalent bond formed by UV crosslinking can accurately fix the binding site of RNA and RBP, the binding site of RBP on RNA can be accurately determined by high-throughput sequencing and bioinformatics analysis, and even the interaction between RBP and RNA can be distinguished. This high-resolution detection ability makes CLIP-Seq play an irreplaceable role in studying the precise mechanism of RNA-protein interaction, such as the location of binding sites between miRNA and mRNA, and the analysis of binding patterns between transcription factors and RNA.
Although RIP-Seq is not as good as CLIP-Seq in resolution, it is sensitive to capture weak interactions. Because RIP-Seq does not cross-link with UV, it can retain some weak but biologically significant RNA-protein interactions in cells under relatively mild experimental conditions. These weak interactions may play an important role in the physiological regulation of cells, such as participating in RNA processing, transportation and translation regulation. Through RIP-Seq, we can detect the existence of these weak interactions and analyze their changes in different physiological and pathological conditions, which provides important information for further understanding the regulatory network of RNA-protein interaction.
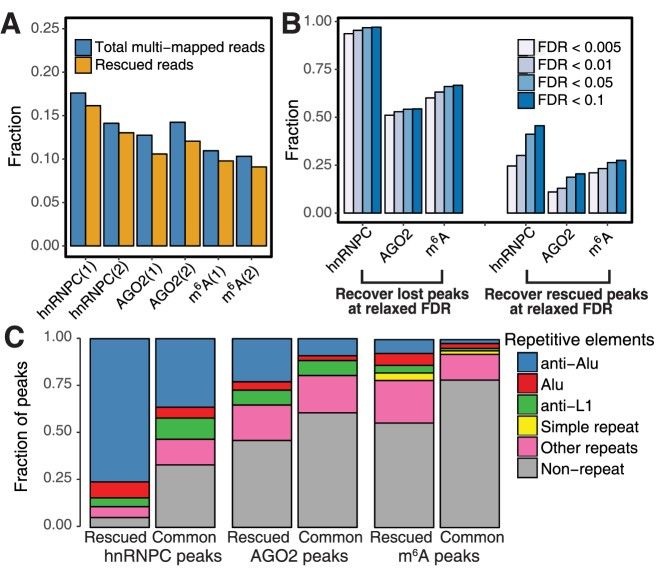 Summary statistics of CLAM results on three CLIP-seq/RIP-seq datasets (Zhang et al., 2017)
Summary statistics of CLAM results on three CLIP-seq/RIP-seq datasets (Zhang et al., 2017)
Dynamic Interaction vs. Stable Interaction
RIP-Seq technology is based on the principle of antigen-antibody specific binding and captures RNA-protein complex under relatively mild conditions. This relatively mild experimental condition enables RIP-Seq to preserve the physiological state of RNA-protein interaction in cells, and is especially suitable for studying dynamic and instantaneous interactions. After cells are stimulated by external stimuli (such as hormones, growth factors, etc.), the binding between RBP and RNA may change rapidly, forming an instantaneous interaction and participating in the rapid response process of cells. RIP-Seq can capture these instantaneous changes, and reveal the dynamic regulation mechanism of RNA-protein interaction in cell physiological process by analyzing the binding spectrum of RBP at different time points.
CLIP-Seq technology uses ultraviolet radiation to covalently cross-link RNA-protein during the experiment to form a stable complex. This strong cross-linking property enables CLIP-Seq to effectively capture RNA-protein interactions that exist stably in cells. CLIP-Seq has unique advantages for some RBP-RNA complexes that are stably bound in cells for a long time. It can identify the direct binding site of RBP and RNA more accurately, and is especially suitable for studying RNA-protein interaction with stable binding pattern.
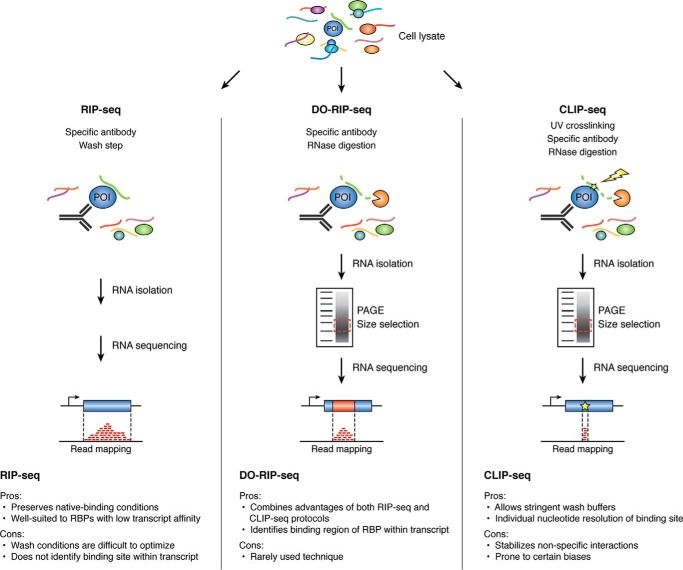 Comparative overview, advantages and disadvantages of pulldown methods for studying RNA–protein interactions (Moore et al., 2019)
Comparative overview, advantages and disadvantages of pulldown methods for studying RNA–protein interactions (Moore et al., 2019)
Low Abundance RNA Detection Ability
The content of low-abundance RNA in cells is very small, and its detection has always been a difficult problem in life science research. CLIP-Seq can covalently cross-link low-abundance RNA with RBP due to its ultraviolet cross-linking characteristics, effectively enriching rare targets. In the cross-linking process, even RNA with extremely low content can be immobilized and detected in the subsequent immunoprecipitation and sequencing process as long as there is interaction with RBP. This characteristic makes CLIP-Seq have unique advantages in the study of low-abundance RNA.
RIP-Seq captures RNA-protein complex under relatively mild conditions, and its ability to detect low-abundance RNA is relatively weak. Due to the low content of RNA in cells, the binding signal with RBP may be weak, which is easy to be missed in RIP-Seq experiment. Especially when the binding affinity between low-abundance RNA and RBP is low, it is difficult to effectively enrich in the process of immunoprecipitation, which leads to the inability to accurately detect the interaction between them.
Comparison between RIP-Seq and CLIP-Seq
| Aspect | RIP-Seq | CLIP-Seq |
|---|---|---|
| Crosslinking | None (natural interactions) | UV-induced covalent crosslinking |
| Resolution | Genomic region (100–300 bp) | Single-nucleotide precision |
| Sensitivity | Better for weak, transient interactions | Better for low-abundance RNAs |
| Antibody Requirement | High specificity (low background) | Tolerates moderate non-specific binding |
| Dynamic vs. Stable RPIs | Ideal for dynamic, stimulus-driven interactions | Excels in stable, constitutive interactions |
How to Choose in Disease Research
In biology research, different biological problems and research purposes have different requirements for technology. Understanding the adaptability of RIP-Seq and CLIP-Seq in different application scenarios is of great significance for accurately analyzing RNA-protein interaction mechanism and promoting biomedical research progress.
Neurodegenerative Diseases
The pathogenesis of neurodegenerative diseases (such as Alzheimer's disease, Parkinson's disease, etc.) is complex, involving abnormal changes in various RNA-protein interactions. CLIP-Seq is widely used in the study of RBP related to neurodegenerative diseases, such as TDP-43.
- TDP-43 is a kind of RBP which plays an important role in neurodegenerative diseases. Its abnormal combination with RNA is closely related to the injury and death of neurons.
- CLIP-Seq can accurately identify the binding site between TDP-43 and RNA, and help researchers to deeply understand the mechanism of TDP-43 in the occurrence and development of neurodegenerative diseases.
- According to CLIP-Seq research, the binding sites of TDP-43 and some specific mRNA changed in the brain tissue of patients with neurodegenerative diseases, which led to the dysfunction of these mRNA, and then affected the normal physiological function of neurons.
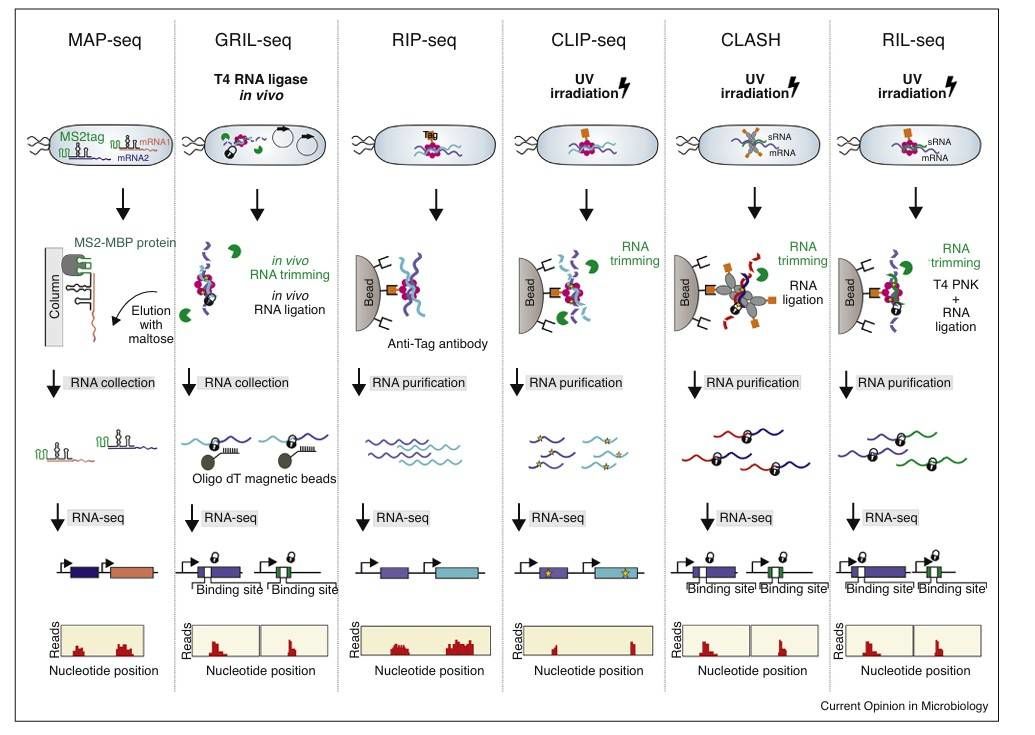 New RNA-seq based methods to decipher regulatory RNA networks (Saliba et al., 2017)
New RNA-seq based methods to decipher regulatory RNA networks (Saliba et al., 2017)
Compared with CLIP-Seq, RIP-Seq has obvious limitations in the study of neurodegenerative diseases. RIP-Seq does not carry out the ultraviolet cross-linking step, so it is difficult to effectively capture protein-RNA aggregates that are unstable and easily dissociated under pathological conditions. In neurodegenerative diseases, abnormal aggregates formed by proteins such as TDP-43 and FUS have complex structures and are easily dissociated under conventional experimental conditions. The relatively mild experimental conditions of RIP-Seq can not stably fix the binding of these aggregates to RNA, which leads to the inability to accurately reveal the interaction between them. Therefore, the application of RIP-Seq is greatly limited in studying the mechanism of abnormal protein aggregation and RNA binding errors in neurodegenerative diseases.
Cancer Research
The occurrence and development of cancer is accompanied by the disorder of RNA metabolism. RIP-Seq has obvious advantages in analyzing the global regulation of RNA metabolism by oncogene RNA binding protein. Take carcinogen RBP LIN28B as an example, it plays a key role in the occurrence and development of various tumors.
- Through RIP-Seq technology, many RNA targets bound by LIN28B can be systematically identified at the whole transcriptome level.
- It is found that after binding to these targets, LIN28B can regulate the stability, processing and translation of RNA, thus reprogramming RNA metabolism globally.
The occurrence of cancer is often accompanied by gene mutations, which may affect the binding site between RBP and RNA, and then change the gene expression pattern. CLIP-Seq is excellent in revealing the effect of carcinogenic mutation on RBP binding site by virtue of its single nucleotide resolution. Take SF3B1 mutation as an example, which is common in many hematological malignancies and solid tumors.
- SF _ 3b1 is a key component of splice, and its mutation will lead to the change of RBP binding site, which will lead to the shift of splice site.
- CLIP-Seq can accurately identify the change of RBP binding site caused by SF3B1 mutation, and clearly show how mutation affects RNA splicing process.
- By analyzing the changes of these sites, we can deeply understand the role of abnormal splicing in the occurrence and development of cancer, and provide theoretical basis for developing cancer treatment drugs against abnormal splicing.
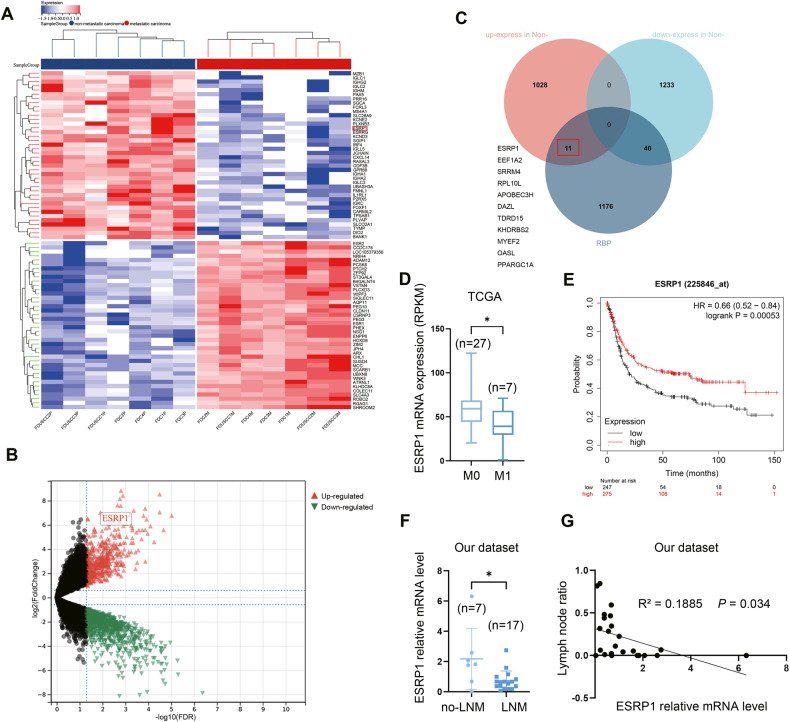 RNA binding protein ESRP1 is downregulated in metastatic gastric cance (Li et al., 2023)
RNA binding protein ESRP1 is downregulated in metastatic gastric cance (Li et al., 2023)
Conclusion
There are many differences in technical principles between RIP-Seq and CLIP-Seq, each with its own advantages and limitations. RIP-Seq is based on natural RNA-protein complex immunoprecipitation, which is suitable for studying dynamic and instantaneous interactions and capturing weak interactions. CLIP-Seq relies on UV-linked immobilization of RNA-protein interaction, which has the advantage of single nucleotide resolution in accurately identifying RNA-protein binding sites.
In the research of RNA-protein interaction, we should fully understand the technical principles of RIP-Seq and CLIP-Seq, reasonably select technical means according to the specific research purposes, sample characteristics and experimental requirements, and combine the two technologies or their variants when necessary, so as to obtain more comprehensive and accurate research results and promote the research in the field of RNA-protein interaction.
References
- Wen J, Parker BJ, Jacobsen A, Krogh A. "MicroRNA transfection and AGO-bound CLIP-seq data sets reveal distinct determinants of miRNA action." RNA. 2011 17(5): 820-34 https://doi.org/10.1261/rna.2387911
- Barra J, Leucci E. "Probing Long Non-coding RNA-Protein Interactions." Front. Mol. Biosci. 2017 (4) https://doi.org/10.3389/fmolb.2017.00045
- Zhang Z, Xing Y. "CLIP-seq analysis of multi-mapped reads discovers novel functional RNA regulatory sites in the human transcriptome." Nucleic Acids Res. 2017 45(16): 9260-9271 https://doi.org/10.1093/nar/gkx646
- Moore KS, 't Hoen PAC. "Computational approaches for the analysis of RNA-protein interactions: A primer for biologists." J Biol Chem. 2019 294(1): 1-9 https://doi.org/10.1074/jbc.rev118.004842
- Saliba AE, C Santos S, Vogel J. "New RNA-seq approaches for the study of bacterial pathogens." Curr Opin Microbiol. 2017 35: 78-87 https://doi.org/10.1016/j.mib.2017.01.001
- Li C, Yin Y., et al. "ESRP1-driven alternative splicing of CLSTN1 inhibits the metastasis of gastric cancer." Cell Death Discov. 2023 9(1): 464 https://doi.org/10.1038/s41420-023-01757-8




